5YA1
 
 | | crystal structure of LsrK-HPr complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
5XYV
 
 | |
5YA0
 
 | | Crystal structure of LsrK and HPr complex | | Descriptor: | Autoinducer-2 kinase, HEXANE-1,6-DIOL, PHOSPHATE ION, ... | | Authors: | Ryu, K.S, Ha, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
3M5N
 
 | |
3M5O
 
 | |
3M5L
 
 | | Crystal structure of HCV NS3/4A protease in complex with ITMN-191 | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3/4A, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M5M
 
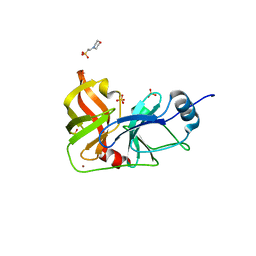 | |
3A2O
 
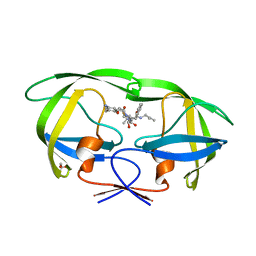 | | Crystal Structure of HIV-1 Protease Complexed with KNI-1689 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(4-amino-2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-5,5-dimethyl-N-(2-methylprop -2-en-1-yl)-1,3-thiazolidine-4-carboxamide, GLYCEROL, PROTEASE | | Authors: | Adachi, M, Tamada, T, Hidaka, K, Kimura, T, Kiso, Y, Kuroki, R. | | Deposit date: | 2009-05-26 | | Release date: | 2010-03-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Small-sized human immunodeficiency virus type-1 protease inhibitors containing allophenylnorstatine to explore the S2' pocket.
J.Med.Chem., 52, 2009
|
|
3CB8
 
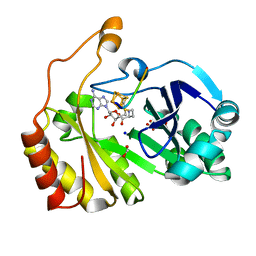 | |
3C8F
 
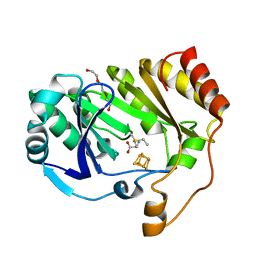 | |
1AGA
 
 | | THE AGAROSE DOUBLE HELIX AND ITS FUNCTION IN AGAROSE GEL STRUCTURE | | Descriptor: | beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | The agarose double helix and its function in agarose gel structure.
J.Mol.Biol., 90, 1974
|
|
1CAR
 
 | | I-CARRAGEENAN. MOLECULAR STRUCTURE AND PACKING OF POLYSACCHARIDE DOUBLE HELICES IN ORIENTED FIBRES OF DIVALENT CATION SALTS | | Descriptor: | 4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Iota-carrageenan: molecular structure and packing of polysaccharide double helices in oriented fibres of divalent cation salts.
J.Mol.Biol., 90, 1974
|
|
1CLL
 
 | |
4D79
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with ATP at 1.768 Angstroem resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
4D7A
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with AMP at 1.801 Angstroem resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
5LQ4
 
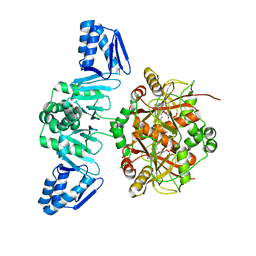 | | The Structure of ThcOx, the First Oxidase Protein from the Cyanobactin Pathways | | Descriptor: | CyaGox, FLAVIN MONONUCLEOTIDE | | Authors: | Bent, A.F, Wagner, A, Naismith, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4DXB
 
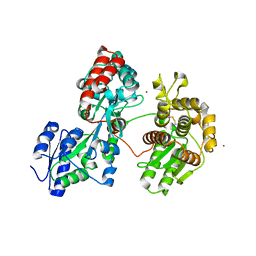 | |
4DXC
 
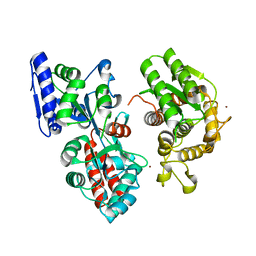 | |
4E90
 
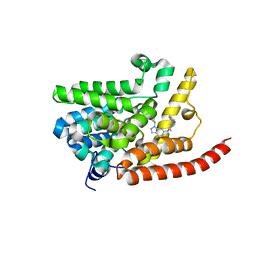 | | Human phosphodiesterase 9 in complex with inhibitors | | Descriptor: | 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-03-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
1HIO
 
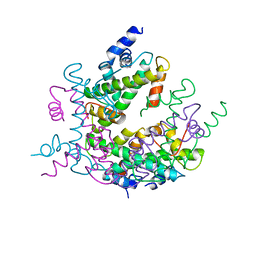 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN, ALPHA CARBONS ONLY | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1991-09-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4G2J
 
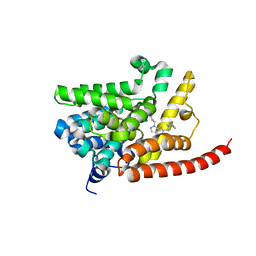 | | Human pde9 in complex with selective compound | | Descriptor: | 1-cyclopentyl-6-[(1R)-1-(3-phenoxyazetidin-1-yl)ethyl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
4G2L
 
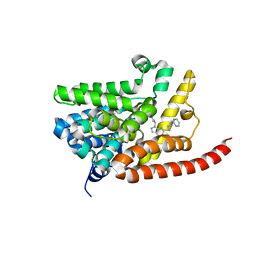 | | Human PDE9 in complex with selective compound | | Descriptor: | 1-cyclopentyl-6-{(1R)-1-[3-(pyrimidin-2-yl)azetidin-1-yl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
7L06
 
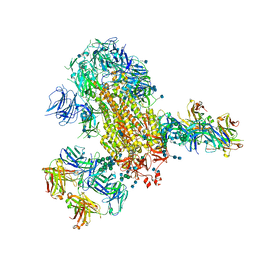 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7L02
 
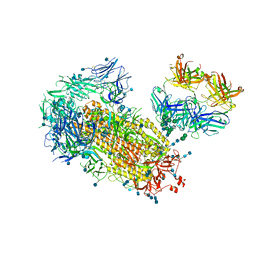 | | Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 heavy chain, ... | | Authors: | Manne, K, Henderson, R, Acharya, P. | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
