3S2U
 
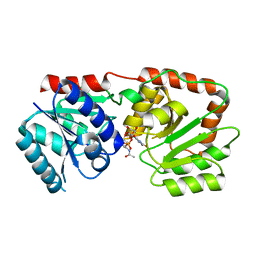 | | Crystal structure of the Pseudomonas aeruginosa MurG:UDP-GlcNAc substrate complex | | Descriptor: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Brown, K, Vial, S.C.M, Dedi, N, Westcott, J, Scally, S, Bugg, T.D.H, Charlton, P.A, Cheetham, G.M.T. | | Deposit date: | 2011-05-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa MurG: UDP-GlcNAc Substrate Complex.
Protein Pept.Lett., 20, 2013
|
|
1SM2
 
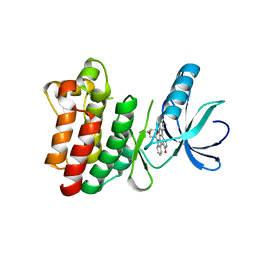 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
2EVA
 
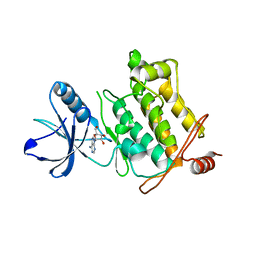 | | Structural Basis for the Interaction of TAK1 Kinase with its Activating Protein TAB1 | | Descriptor: | ADENOSINE, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Brown, K, Vial, S.C, Dedi, N, Long, J.M, Dunster, N.J, Cheetham, G.M. | | Deposit date: | 2005-10-31 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the interaction of TAK1 kinase with its activating protein TAB1
J.Mol.Biol., 354, 2005
|
|
1SNU
 
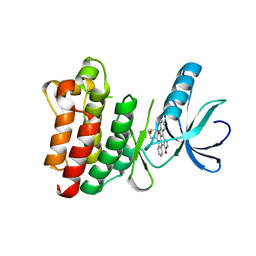 | | CRYSTAL STRUCTURE OF THE UNPHOSPHORYLATED INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
1SNX
 
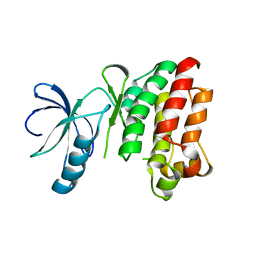 | | CRYSTAL STRUCTURE OF APO INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
7ZBN
 
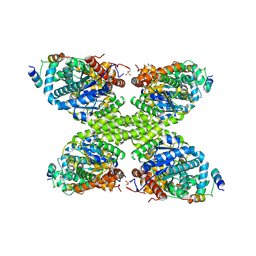 | |
