2FYI
 
 | | Crystal Structure of the Cofactor-Binding Domain of the Cbl Transcriptional Regulator | | Descriptor: | HTH-type transcriptional regulator cbl | | Authors: | Stec, E, Neumann, P, Wilkinson, A.J, Brzozowski, A.M, Bujacz, G.D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Sulphate Starvation Response in E. coli: Crystal Structure and Mutational Analysis of the Cofactor-binding Domain of the Cbl Transcriptional Regulator.
J.Mol.Biol., 364, 2006
|
|
1DTE
 
 | |
1DT3
 
 | |
1DT5
 
 | |
1DU4
 
 | |
1EIN
 
 | |
7XTP
 
 | | eIF4E in Complex with a Disulphide-Free Autonomous VH Domain | | Descriptor: | Eukaryotic translation initiation factor 4E, VH-S4ss, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Brown, C.J, Frosi, Y, Jiang, S, Lin, Y.C. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex.
Nat Commun, 13, 2022
|
|
7BWE
 
 | | Consensus Chitin binding domain | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7BWO
 
 | | Consensus chitin binding protein | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7D8B
 
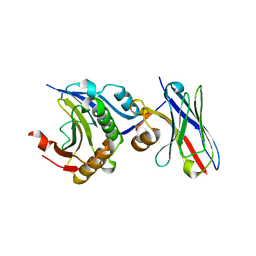 | | Engineering Disulphide-Free Autonomous Antibody VH Domains to modulate intracellular pathways | | Descriptor: | Eukaryotic translation initiation factor 4E, VH-S4 | | Authors: | Frosi, Y, Lin, Y.C, Jiang, S, Brown, C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex.
Nat Commun, 13, 2022
|
|
6Q38
 
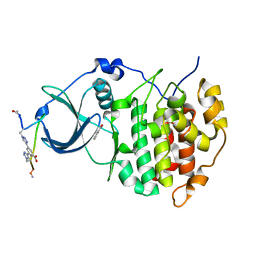 | | The Crystal structure of CK2a bound to P1-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, BENZOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
6Q4Q
 
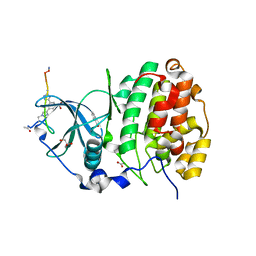 | | The Crystal structure of CK2a bound to P2-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, ACETATE ION, BENZOIC ACID, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
7F07
 
 | | Autonomous VH domain that interacts with eIF4E at the Capped mRNA Binding site. | | Descriptor: | Eukaryotic translation initiation factor 4E, VH domain (VH-DiFCAP-01) | | Authors: | Brown, C.J, Frosi, Y, Ng, S, Lin, Y.C. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
7EZW
 
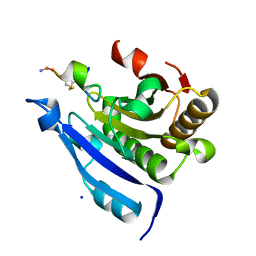 | | Cyclic Peptide that Interacts with the eIF4E Capped-mRNA Binding Site | | Descriptor: | ALA-CYS-GLU-MET-GLY-PHE-PHE-GLN-ASP-CYS-GLY, Eukaryotic translation initiation factor 4E, SODIUM ION | | Authors: | Brown, C.J, Ng, S, Frosi, Y. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
6GK0
 
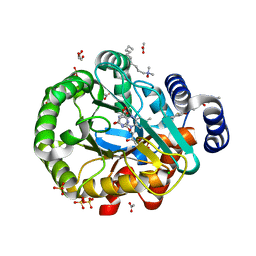 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH CLASS III HISTONE DEACETYLASE INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 4-~{tert}-butyl-~{N}-[[4-[5-(dimethylamino)pentanoylamino]phenyl]carbamothioyl]benzamide, ACETIC ACID, ... | | Authors: | Hakansson, M, Ladds, M.J.G.W, Walse, B, Lain, S. | | Deposit date: | 2018-05-17 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J.Biol.Chem., 295, 2020
|
|
5XWR
 
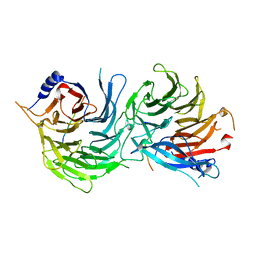 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6IDK
 
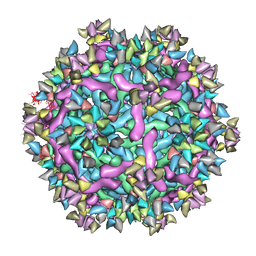 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class I particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
6IDI
 
 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 8.0. | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
6IDL
 
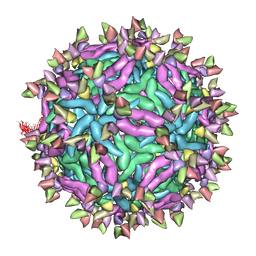 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class II particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
1AKU
 
 | | D95A HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKQ
 
 | | D95A OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-03-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1AKV
 
 | | D95A SEMIQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic investigation of the role of aspartate 95 in the modulation of the redox potentials of Desulfovibrio vulgaris flavodoxin.
Biochemistry, 41, 2002
|
|
1C7F
 
 | | D95E OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | McCarthy, A, Walsh, M, Higgins, T, D'Arcy, D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Investigation of the Role of Aspartate 95 in the Modulation of the Redox Potentials Of Desulfovibrio Vulgaris Flavodoxin
Biochemistry, 41, 2002
|
|
1C7E
 
 | | D95E HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | McCarthy, A, Walsh, M, Higgins, T, D'Arcy, D. | | Deposit date: | 2000-02-16 | | Release date: | 2000-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic Investigation of the Role of Aspartate 95 in the Modulation of the Redox Potentials Of Desulfovibrio Vulgaris Flavodoxin
Biochemistry, 41, 2002
|
|
6LEK
 
 | | Tertiary structure of Barnacle cement protein MrCP20 | | Descriptor: | Cement protein-20k | | Authors: | Mohanram, H. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of Megabalanus rosa Cement Protein 20 revealed by multi-dimensional NMR and molecular dynamics simulations.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 374, 2019
|
|
