5J3Q
 
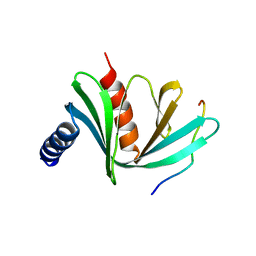 | | Crystal structure of S. pombe Dcp1:Edc1 mRNA decapping complex | | Descriptor: | Edc1, mRNA-decapping enzyme subunit 1 | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5J3T
 
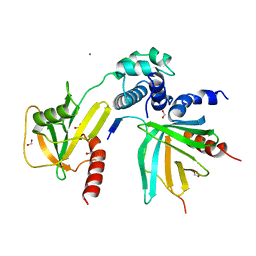 | | Crystal structure of S. pombe Dcp2:Dcp1:Edc1 mRNA decapping complex | | Descriptor: | Edc1, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5J3Y
 
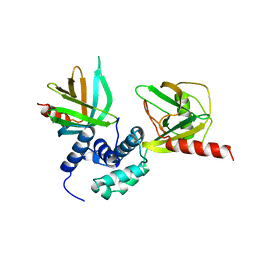 | | Crystal structure of S. pombe Dcp2:Dcp1 mRNA decapping complex | | Descriptor: | mRNA decapping complex subunit 2, mRNA-decapping enzyme subunit 1 | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2Y92
 
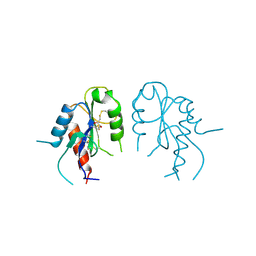 | | Crystal structure of MAL adaptor protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TOLL/INTERLEUKIN-1 RECEPTOR DOMAIN-CONTAINING ADAPTER PROTEIN, | | Authors: | Valkov, E, Stamp, A, Martin, J.L, Kobe, B. | | Deposit date: | 2011-02-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Toll-Like Receptor Adaptor Mal/Tirap Reveals the Molecular Basis for Signal Transduction and Disease Protection.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3DLR
 
 | |
5C5W
 
 | | 1.25 A resolution structure of an RNA 20-mer | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*AP*GP*UP*UP*CP*AP*AP*UP*UP*CP*UP*AP*GP*CP*G)-3') | | Authors: | Stewart, M, Valkov, E. | | Deposit date: | 2015-06-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6FNY
 
 | | CRYSTAL STRUCTURE OF A CHOLINE SULFATASE FROM SINORHIZOBIUM MELLILOTI | | Descriptor: | CALCIUM ION, Choline-sulfatase | | Authors: | Valkov, E, Van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and Mechanistic Analysis of the Choline Sulfatase from Sinorhizobium melliloti: A Class I Sulfatase Specific for an Alkyl Sulfate Ester.
J. Mol. Biol., 430, 2018
|
|
4WPX
 
 | | Chaetomium theromophilum TREX2 CID domain complex | | Descriptor: | Cell division control protein 31-like protein, Putative SAC3 family protein, Putative uncharacterized protein | | Authors: | Valkov, E, Stewart, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Structure, 23, 2015
|
|
4AF0
 
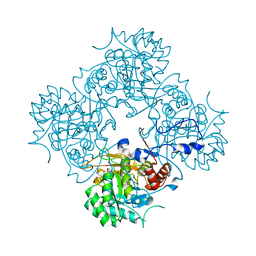 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
4UPK
 
 | |
8FY3
 
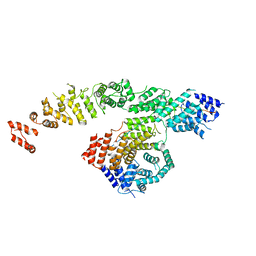 | | Structure of NOT1:NOT10:NOT11 module of the human CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Pekovic, F, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
8FY4
 
 | | Structure of NOT1:NOT10:NOT11 module of the chicken CCR4-NOT complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Lea, S.M, Deme, J.C, Raisch, T, Levdansky, Y, Valkov, E. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and assembly of the NOT10:11 module of the CCR4-NOT complex.
Commun Biol, 6, 2023
|
|
5FU7
 
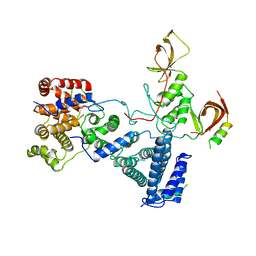 | | drosophila nanos NBR peptide bound to the NOT module of the human CCR4-NOT complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 2, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 3, ... | | Authors: | Raisch, T, Bhandari, D, Sabath, K, Helms, S, Valkov, E, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct Modes of Recruitment of the Ccr4-not Complex by Drosophila and Vertebrate Nanos
Embo J., 35, 2016
|
|
5FU6
 
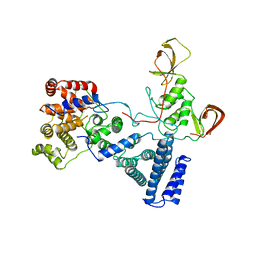 | | NOT module of the human CCR4-NOT complex (Crystallization mutant) | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 2, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 3 | | Authors: | Raisch, T, Bhandari, D, Sabath, K, Helms, S, Valkov, E, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distinct Modes of Recruitment of the Ccr4-not Complex by Drosophila and Vertebrate Nanos
Embo J., 35, 2016
|
|
6S8R
 
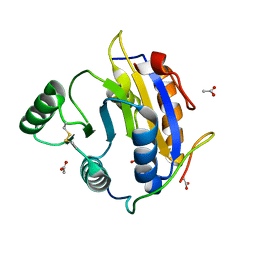 | | D. melanogaster RNA helicase Me31B in complex with GIGYF | | Descriptor: | ACETATE ION, ATP-dependent RNA helicase me31b, GIGYF family protein CG11148 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression.
Genes Dev., 33, 2019
|
|
6S8S
 
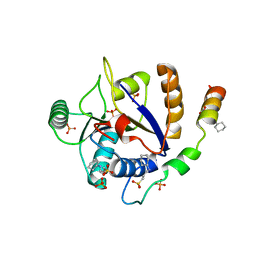 | |
4LJ0
 
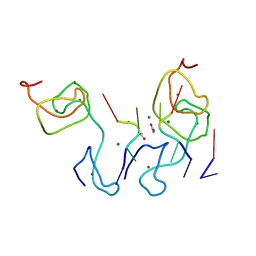 | | Nab2 Zn fingers complexed with polyadenosine | | Descriptor: | ACETATE ION, MAGNESIUM ION, Nab2, ... | | Authors: | Stewart, M, Kuhlmann, S.I, Valkov, E. | | Deposit date: | 2013-07-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers.
Nucleic Acids Res., 42, 2014
|
|
3ZLG
 
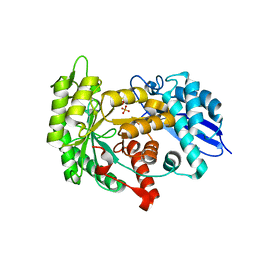 | | Structure of group A Streptococcal enolase K362A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3ZLH
 
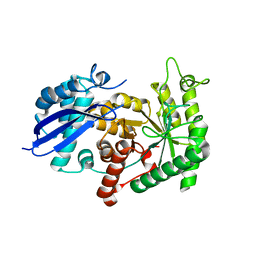 | | Structure of group A Streptococcal enolase | | Descriptor: | ENOLASE | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
3ZTT
 
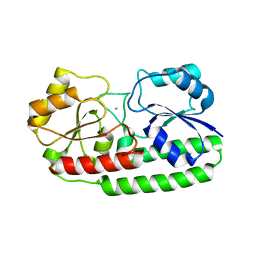 | | Crystal structure of pneumococcal surface antigen PsaA with manganese | | Descriptor: | MANGANESE (II) ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | McDevitt, C.A, Ogunniyi, A.D, Valkov, E, Lawrence, M.C, Kobe, B, McEwan, A.G, Paton, J.C. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular mechanism for bacterial susceptibility to zinc.
PLoS Pathog., 7, 2011
|
|
5T47
 
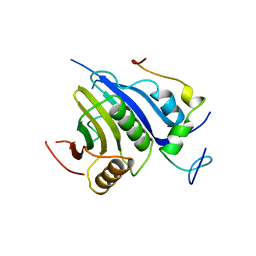 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, isoform A, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
5T46
 
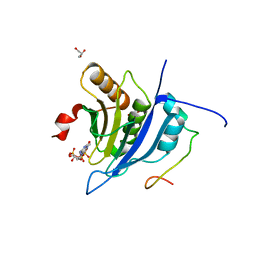 | | Crystal structure of the human eIF4E-eIF4G complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
5T48
 
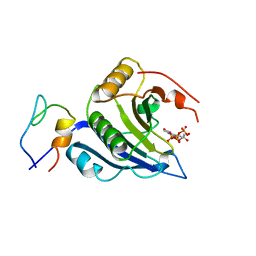 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex without lateral contact | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
3ZLF
 
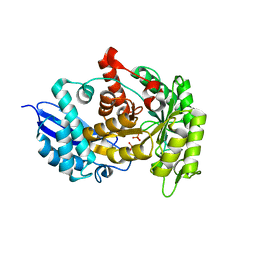 | | Structure of group A Streptococcal enolase K312A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
5NVM
 
 | |
