2WS7
 
 | | Semi-synthetic analogue of human insulin ProB26-DTI | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WS0
 
 | | Semi-synthetic analogue of human insulin NMeAlaB26-insulin at pH 7.5 | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WS6
 
 | | Semi-synthetic analogue of human insulin NMeTyrB26-insulin in hexamer form | | Descriptor: | CHLORIDE ION, GLYCEROL, INSULIN A CHAIN, ... | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WRX
 
 | | Semi-synthetic analogue of human insulin NMeAlaB26-insulin at pH 3.0 | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SODIUM ION | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6Y5S
 
 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6Y5T
 
 | | Crystal structure of savinase at room temperature | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
4TGL
 
 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
3D55
 
 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
3CTO
 
 | | Crystal Structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
6H75
 
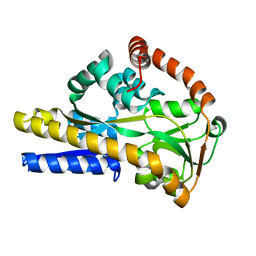 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
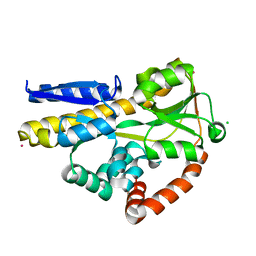 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
5LWH
 
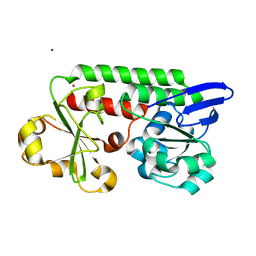 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
1DTE
 
 | |
1DT3
 
 | |
1DT5
 
 | |
1DYO
 
 | | Xylan-Binding Domain from CBM 22, formally x6b domain | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Davies, G.J, Charnock, S.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2000-02-03 | | Release date: | 2000-07-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X6 Thermostabilising Domains of Xylanases are Carbohydrate Binding Modules: Structure and Biochemistry of the Clostridium Thermocellum X6B Domain
Biochemistry, 39, 2000
|
|
1DU4
 
 | |
1EIN
 
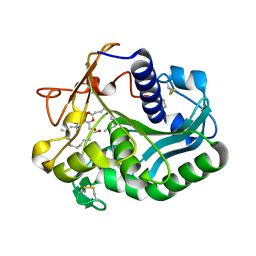 | |
6FF3
 
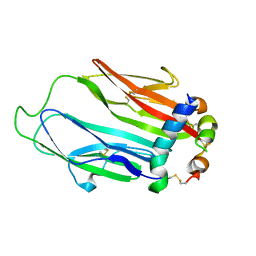 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L1 with Human IGF-I | | Descriptor: | Insulin-like growth factor I, Neural/ectodermal development factor IMP-L2 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D, Viola, C.M. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
6FEY
 
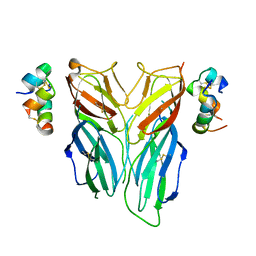 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L2 with Drosophila DILP5 insulin | | Descriptor: | Neural/ectodermal development factor IMP-L2, Probable insulin-like peptide 5 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
6S5Y
 
 | |
6S5Z
 
 | | Structure of Rib R28N from Streptococcus pyogenes | | Descriptor: | SODIUM ION, Surface protein R28 | | Authors: | Whelan, F, Griffiths, S.C, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S5W
 
 | | Structure of Rib domain 'Rib Long' from Lactobacillus acidophilus | | Descriptor: | SODIUM ION, SULFATE ION, Surface protein | | Authors: | Griffiths, S.C, Cooper, R.E.M, Whelan, F, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6G1M
 
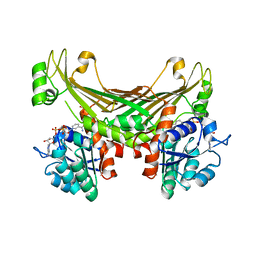 | | Amine Dehydrogenase from Petrotoga mobilis; open and closed form | | Descriptor: | Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Beloti, L, Frese, A, Mayol, O, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
