6DJJ
 
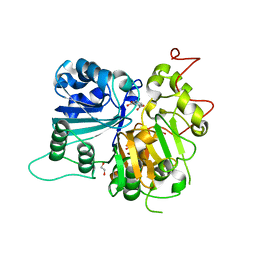 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DIH
 
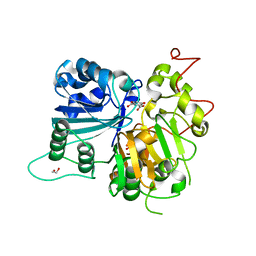 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJG
 
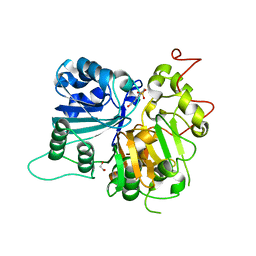 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ503 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-sulfoquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
1XKP
 
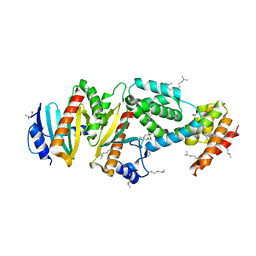 | | Crystal structure of the virulence factor YopN in complex with its heterodimeric chaperone SycN-YscB | | Descriptor: | Chaperone protein sycN, Chaperone protein yscB, putative membrane-bound Yop targeting protein YopN | | Authors: | Schubot, F.D, Jackson, M.W, Penrose, K.J, Cherry, S, Tropea, J.E, Plano, G.V, Waugh, D.S. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis.
J.Mol.Biol., 346, 2005
|
|
1XL3
 
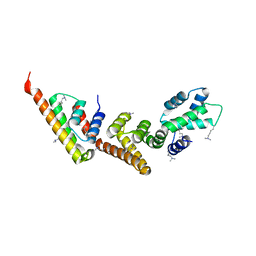 | | Complex structure of Y.pestis virulence Factors YopN and TyeA | | Descriptor: | Secretion control protein, protein type A | | Authors: | Schubot, F.D, Jackson, M.W, Penrose, K.J, Cherry, S, Tropea, J.E, Plano, G.V, Waugh, D.S. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis.
J.Mol.Biol., 346, 2005
|
|
1Z21
 
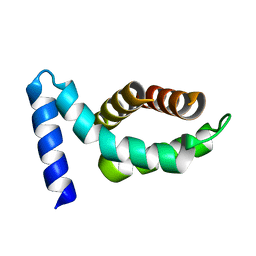 | | Crystal structure of the core domain of Yersinia pestis virulence factor YopR | | Descriptor: | Yop proteins translocation protein H | | Authors: | Schubot, F.D, Cherry, S, Tropea, J.E, Austin, B.P, Waugh, D.S. | | Deposit date: | 2005-03-07 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of the protease-resistant core domain of Yersinia pestis virulence factor YopR.
Protein Sci., 14, 2005
|
|
1YYK
 
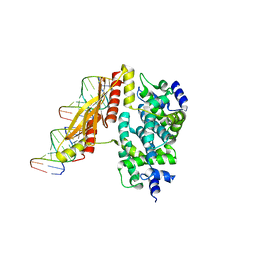 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1Z0E
 
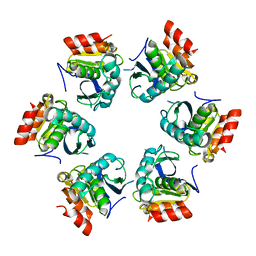 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1YYO
 
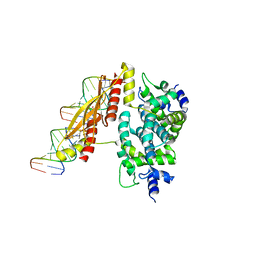 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YZ9
 
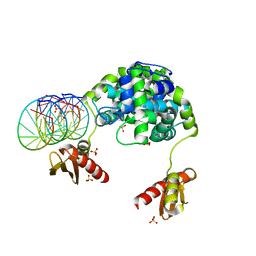 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYW
 
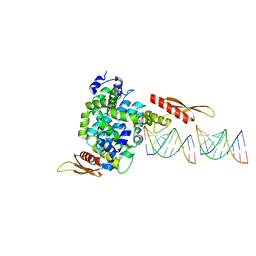 | | Crystal structure of RNase III from Aquifex aeolicus complexed with double stranded RNA at 2.8-Angstrom Resolution | | Descriptor: | 5'-R(*AP*AP*AP*UP*AP*UP*AP*UP*AP*UP*UP*U)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1Z0C
 
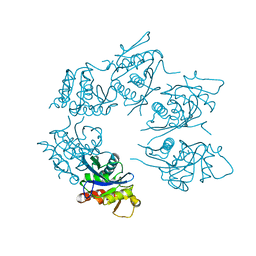 | | Crystal Structure of A. fulgidus Lon proteolytic domain D508A mutant | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0G
 
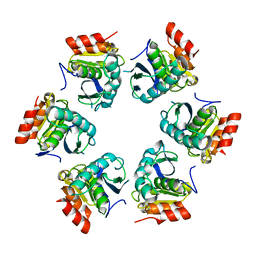 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0W
 
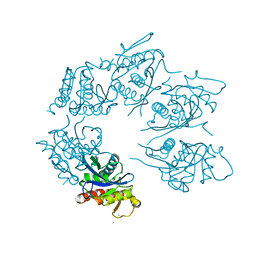 | | Crystal Structure of A. fulgidus Lon proteolytic domain at 1.2A resolution | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0B
 
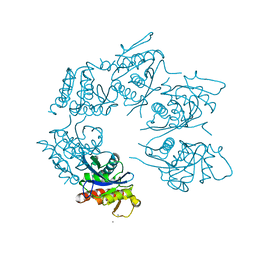 | | Crystal Structure of A. fulgidus Lon proteolytic domain E506A mutant | | Descriptor: | CALCIUM ION, Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
2IF5
 
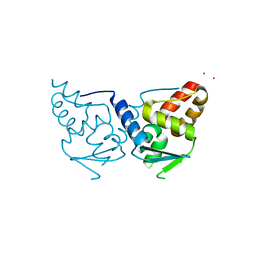 | | Structure of the POZ domain of human LRF, a master regulator of oncogenesis | | Descriptor: | PRASEODYMIUM ION, Zinc finger and BTB domain-containing protein 7A | | Authors: | Schubot, F.D, Waugh, D.S, Tropea, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the POZ domain of human LRF, a master regulator of oncogenesis.
Biochem.Biophys.Res.Commun., 351, 2006
|
|
3R9X
 
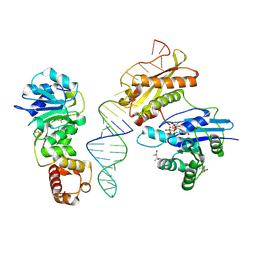 | | Crystal structure of Era in complex with MgGDPNP, nucleotides 1506-1542 of 16S ribosomal RNA, and KsgA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, GTPase Era, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2011-03-26 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3R9W
 
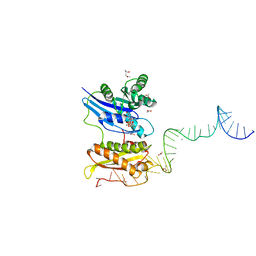 | |
4PZV
 
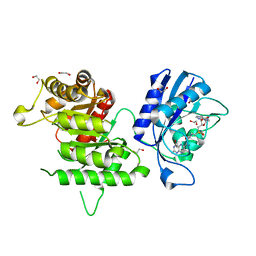 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with bisubstrate analog HPPK inhibitor J1D | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK-DHPS from the biowarfare agent Francisella tularensis.
Febs J., 281, 2014
|
|
1RC7
 
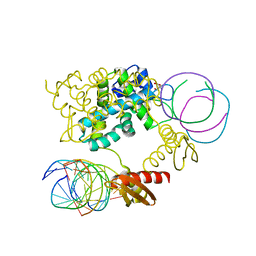 | | Crystal structure of RNase III Mutant E110K from Aquifex Aeolicus complexed with ds-RNA at 2.15 Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*GP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3', Ribonuclease III | | Authors: | Blaszczyk, J, Gan, J, Ji, X. | | Deposit date: | 2003-11-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Noncatalytic Assembly of Ribonuclease III with Double-Stranded RNA.
Structure, 12, 2004
|
|
1RC5
 
 | |
3T92
 
 | | Crystal structure of the Taz2:C/EBPepsilon-TAD chimera protein | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ACETONE, HISTONE ACETYLTRANSFERASE P300 TAZ2-CCAAT/ENHANCER-BINDING PROTEIN EPSILON, ... | | Authors: | Bhaumik, P, Maria, M. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into interactions of C/EBP transcriptional activators with the Taz2 domain of p300.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
6PC1
 
 | | Crystal structure of Helicobacter pylori PPX/GppA (E143A) in complex with ppGpp | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PC0
 
 | | Crystal structure of Helicobacter pylori PPX/GppA | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, D-MALATE, ... | | Authors: | Song, H, Wang, C, Shaw, G.X, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
6PBZ
 
 | | Crystal structure of Escherichia coli GppA | | Descriptor: | CHLORIDE ION, Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase | | Authors: | Song, H, Shaw, G.X, Wang, C, Ji, X. | | Deposit date: | 2019-06-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori.
Febs J., 287, 2020
|
|
