4A56
 
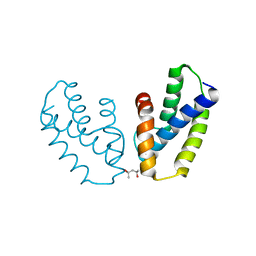 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | Authors: | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
2WCQ
 
 | | Crystal Structure of Tip-Alpha N34 (HP0596) from Helicobacter pylori at pH4 | | Descriptor: | CITRIC ACID, TNF-ALPHA INDUCER PROTEIN | | Authors: | Tosi, T, Cioci, G, Jouravleva, K, Dian, C, Terradot, L. | | Deposit date: | 2009-03-13 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Tumor Necrosis Factor Alpha Inducing Protein Tipalpha: A Novel Virulence Factor from Helicobacter Pylori.
FEBS Lett., 583, 2009
|
|
2WCR
 
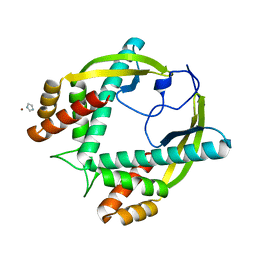 | | Crystal Structure of Tip-Alpha N34 (HP0596) from Helicobacter pylori at pH8 | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, TNF-ALPHA INDUCER PROTEIN | | Authors: | Tosi, T, Cioci, G, Jouravleva, K, Dian, C, Terradot, L. | | Deposit date: | 2009-03-13 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the Tumor Necrosis Factor Alpha Inducing Protein Tipalpha: A Novel Virulence Factor from Helicobacter Pylori.
FEBS Lett., 583, 2009
|
|
6GYZ
 
 | |
6HUW
 
 | |
6GYX
 
 | |
6GYW
 
 | |
6GYY
 
 | |
5IIP
 
 | | Staphylococcus aureus OpuCA | | Descriptor: | Glycine betaine/carnitine/choline ABC transporter%2C ATP-binding protein%2C putative | | Authors: | Tosi, T, Campeotto, I, Freemont, P.S, Grundling, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus.
Sci.Signal., 9, 2016
|
|
6EOW
 
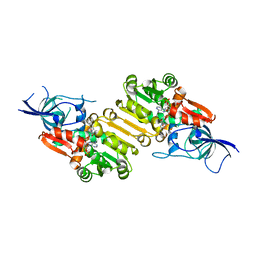 | |
4G0H
 
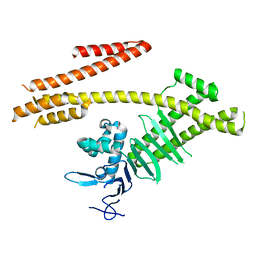 | | Crystal structure of the N-terminal domain of Helicobacter pylori CagA protein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Kaplan-Turkoz, B, Remaut, H, Dian, C, Louche, A, Terradot, L. | | Deposit date: | 2012-07-09 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Helicobacter pylori oncoprotein CagA interaction with beta 1 integrin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7OML
 
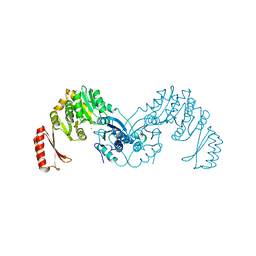 | |
7OLH
 
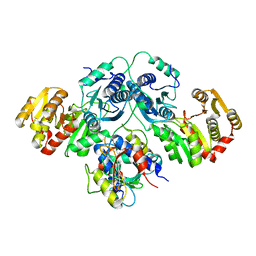 | |
7OJR
 
 | |
7OJS
 
 | |
