2KGQ
 
 | | Refined solution structure of des-pyro Glu brazzein | | Descriptor: | Brazzein | | Authors: | Cornilescu, C.C, Tonelli, M, DeRider, M.L, Markley, J.L, Assadi-Porter, F.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of des-pyro Glu brazzein
To be Published
|
|
2KMW
 
 | |
2KNH
 
 | | The Solution structure of the eTAFH domain of AML1-ETO complexed with HEB peptide | | Descriptor: | Protein CBFA2T1, Transcription factor 12 | | Authors: | Park, S, Cierpicki, T, Tonelli, M, Bushweller, J.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity.
Blood, 113, 2009
|
|
2KQK
 
 | | Solution structure of apo-IscU(D39A) | | Descriptor: | NifU-like protein | | Authors: | Kim, J.H, Fuzery, A.K, Tonelli, M, Vickery, L.E, Markley, J.L. | | Deposit date: | 2009-11-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure and Determinants of Stability of the Iron-Sulfur Cluster Scaffold Protein IscU from Escherichia coli.
Biochemistry, 51, 2012
|
|
2KSI
 
 | | Solution NMR structure of Sterol Carrier Protein - 2 from Aedes aegypti (AeSCP-2) complex with C16 fatty acid (palmitate) | | Descriptor: | PALMITIC ACID, Sterol carrier protein 2 | | Authors: | Singarapu, K, Radek, J.T, Tonelli, M, Lan, Q, Markley, J.L. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Sterol Carrier Protein - 2 from Aedes aegypti (AeSCP-2) complex with C16 fatty acid (palmitate)
To be Published
|
|
2KSH
 
 | |
2L9Q
 
 | |
2L94
 
 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
2L4X
 
 | | Solution Structure of apo-IscU(WT) | | Descriptor: | Iron-sulfur cluster assembly scaffold protein | | Authors: | Kim, J.H, Tonelli, M, Markley, J.L. | | Deposit date: | 2010-10-19 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure and Determinants of Stability of the Iron-Sulfur Cluster Scaffold Protein IscU from Escherichia coli.
Biochemistry, 51, 2012
|
|
2LE4
 
 | | Solution structure of the HMG box DNA-binding domain of human stem cell transcription factor Sox2 | | Descriptor: | Transcription factor SOX-2 | | Authors: | Sahu, S.C, Markley, J.L, Tonelli, M, Bahrami, A, Eghbalnia, H.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG box DNA-binding domain of human stem cell transcription factor Sox2
To be Published
|
|
2M5T
 
 | | Solution structure of the 2A proteinase from a common cold agent, human rhinovirus RV-C02, strain W12 | | Descriptor: | ZINC ION, human rhinovirus 2A proteinase | | Authors: | Lee, W, Frederick, R, Tonelli, M, Troupis, A.T, Reinin, N, Suchy, F.P, Moyer, K, Watters, K, Aceti, D, Palmenberg, A.C, Markley, J.L. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 2A Protease from a Common Cold Agent, Human Rhinovirus C2, Strain W12.
Plos One, 9, 2014
|
|
2LY5
 
 | | Refined solution structure of recombinant brazzein | | Descriptor: | Defensin-like protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Tonelli, M, Markley, J.L, Assadi-Porter, F.M. | | Deposit date: | 2012-09-12 | | Release date: | 2013-01-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational change affecting Tyr11 and sweetness loops of brazzein.
Proteins, 81, 2013
|
|
2LY6
 
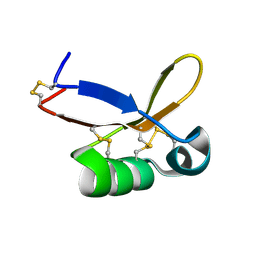 | | Refined solution structure of recombinant brazzein at low temperature | | Descriptor: | Defensin-like protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Tonelli, M, Markley, J.L, Assadi-Porter, F.M. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational change affecting Tyr11 and sweetness loops of brazzein.
Proteins, 81, 2013
|
|
2N66
 
 | |
2N69
 
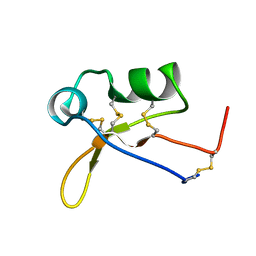 | |
6VEO
 
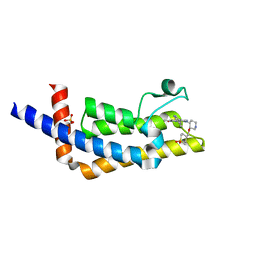 | | ATAD2B bromodomain in complex with 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione (compound 38) | | Descriptor: | 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione, ATPase family AAA domain-containing protein 2B, SULFATE ION | | Authors: | Glass, K.C. | | Deposit date: | 2020-01-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain.
J.Med.Chem., 63, 2020
|
|
4IBK
 
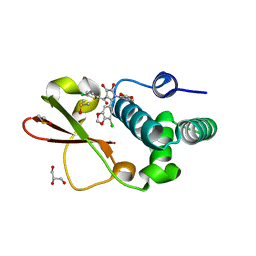 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBC
 
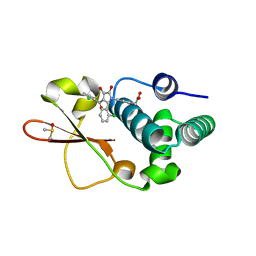 | | Ebola virus VP35 bound to small molecule | | Descriptor: | DIMETHYL SULFOXIDE, Polymerase cofactor VP35, {4-[(2R)-3-(2-chlorobenzoyl)-2-(2-chlorophenyl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
2WC2
 
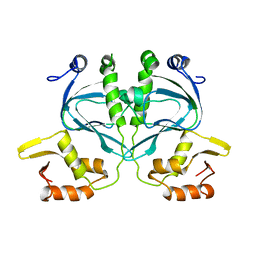 | |
4IBG
 
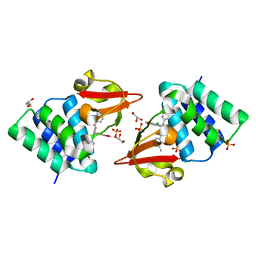 | | Ebola virus VP35 bound to small molecule | | Descriptor: | GLYCEROL, PHOSPHATE ION, Polymerase cofactor VP35, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
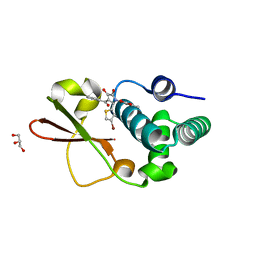 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBI
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBF
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBJ
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
