5X4V
 
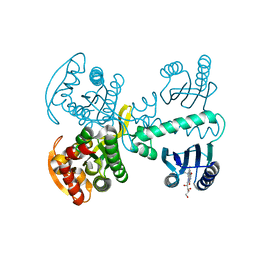 | | Roseoflavin substituted OaPAC | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, Photoactivated adenylyl cyclase | | Authors: | Ohki, M, Park, S.-Y. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of photoactivation of a light-regulated adenylate cyclase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X4U
 
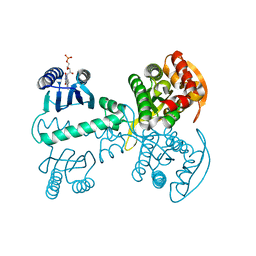 | |
7V73
 
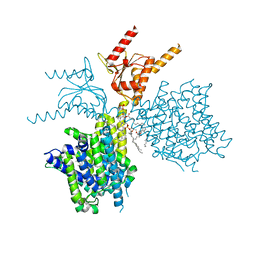 | | Thermostabilized human prestin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V75
 
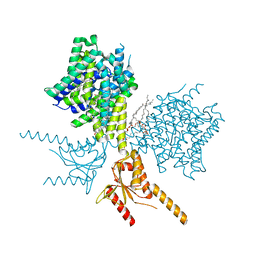 | | Thermostabilized human prestin in complex with salicylate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-HYDROXYBENZOIC ACID, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V74
 
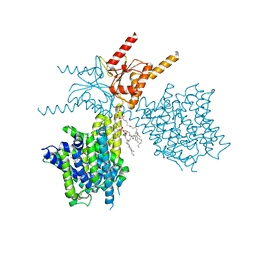 | | Thermostabilized human prestin in complex with sulfate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, SULFATE ION, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
6JYA
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY8
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY7
 
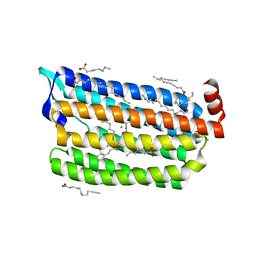 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYD
 
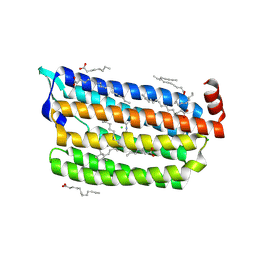 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY9
 
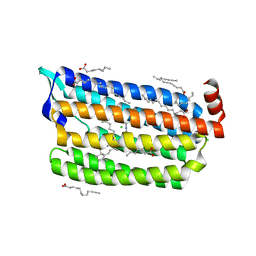 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYB
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6LBH
 
 | |
7V6C
 
 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6B
 
 | | Structure of the Dicer-2-R2D2 heterodimer | | Descriptor: | Dicer-2, isoform A, R2D2 | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
