6A58
 
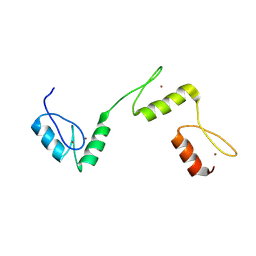 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
6A57
 
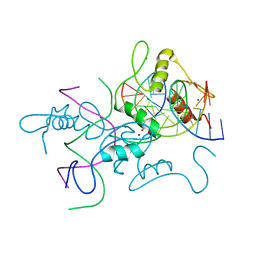 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
6A59
 
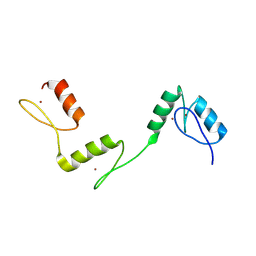 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
6JDR
 
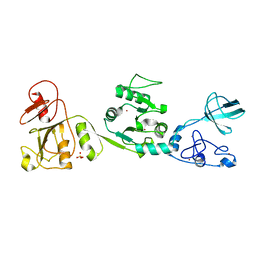 | |
6JDU
 
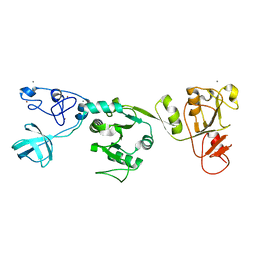 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
2OAE
 
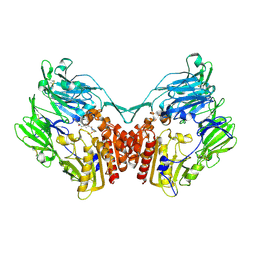 | | Crystal structure of rat dipeptidyl peptidase (DPPIV) with thiazole-based peptide mimetic #31 | | Descriptor: | Dipeptidyl peptidase 4, N-{[(3S,5S)-5-(1,3-THIAZOLIDIN-3-YLCARBONYL)PYRROLIDIN-3-YL]METHYL}-1,3-THIAZOLE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Longenecker, K.L, Shuai, Q, Patel, J, Wiedeman, P. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6JDS
 
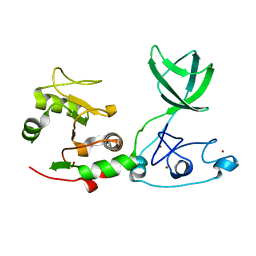 | |
6A3V
 
 | | Complex structure of human 4-1BB and 4-1BBL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Li, Y, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | Deposit date: | 2018-06-17 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
6AEE
 
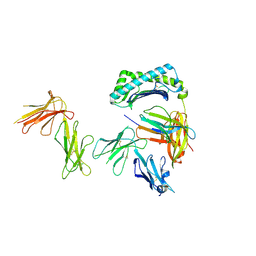 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | Descriptor: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Wang, Q, Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2018-08-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6A3W
 
 | | Complex structure of 4-1BB and utomilumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 9, utomilumab VH, ... | | Authors: | Li, Y, Tan, S, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | Deposit date: | 2018-06-17 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
6AED
 
 | |
7UMG
 
 | | Crystal structure of human CD8aa-MR1-Ac-6-FP complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
J.Exp.Med., 219, 2022
|
|
7VJV
 
 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJS
 
 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
2OAG
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) with pyrrolidine-constrained phenethylamine 29g | | Descriptor: | (3R,4S)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PYRROLIDIN-3-AMINE, Dipeptidyl peptidase 4 | | Authors: | Backes, B.J, Longenecker, K.L, Hamilton, G.L, Stewart, K.D, Lai, C, Kopecka, H. | | Deposit date: | 2006-12-15 | | Release date: | 2007-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6GU1
 
 | |
8XAX
 
 | |
8XAW
 
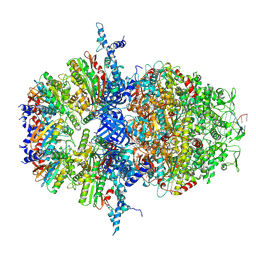 | |
8XAU
 
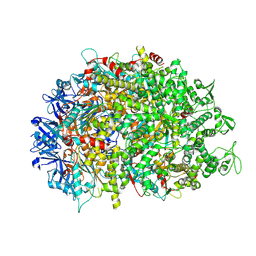 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAY
 
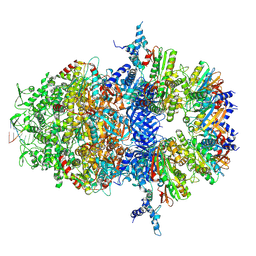 | |
8XAV
 
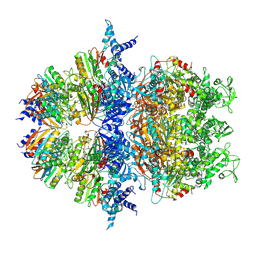 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8J30
 
 | | Crystal structure of ApNGT with Q469A and M218A mutations in complex with UDP-GLC | | Descriptor: | UDP-glucose:protein N-beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Feng, Y, Hao, Z, Guo, Q, Zheng, J, Da, L, Peng, W. | | Deposit date: | 2023-04-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Investigation of the Catalytic Mechanism of a Soluble N-glycosyltransferase Allows Synthesis of N-glycans at Noncanonical Sequons.
Jacs Au, 3, 2023
|
|
7XV9
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV8
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV6
 
 | |
