2MW0
 
 | |
2LMJ
 
 | | Itk-sh3 | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Kristiansen, P, Bie Andersen, T, Huszenicza, Z, Andreotti, A.H, Spurkland, A. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SH3 domains of the Tec family kinase Itk and the Src family kinase Lck compete for adjacent sites on T-cell specific adapter protein
To be Published
|
|
2D3O
 
 | | Structure of Ribosome Binding Domain of the Trigger Factor on the 50S ribosomal subunit from D. radiodurans | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, ... | | Authors: | Schluenzen, F, Wilson, D.N, Hansen, H.A, Tian, P, Harms, J.M, McInnes, S.J, Albrecht, R, Buerger, J, Wilbanks, S.M, Fucini, P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Binding Mode of the Trigger Factor on the Ribosome: Implications for Protein Folding and SRP Interaction
Structure, 13, 2005
|
|
8GZB
 
 | | SARS-CoV-2 3CLpro | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-chlorophenyl)-1,3,4-oxadiazole, 3C-like proteinase nsp5 | | Authors: | Wang, F, Cen, Y.X, Tian, P. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nature-inspired catalytic asymmetric rearrangement of cyclopropylcarbinyl cation.
Sci Adv, 9, 2023
|
|
7VA1
 
 | |
6I0Y
 
 | | TnaC-stalled ribosome complex with the titin I27 domain folding close to the ribosomal exit tunnel | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Su, T, Kudva, R, von Heijne, G, Beckmann, R. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Folding pathway of an Ig domain is conserved on and off the ribosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2L7P
 
 | | ASHH2 a CW domain | | Descriptor: | Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Kristiansen, P, Hoppmann, V, Thorstensen, T, Aalen, R.B, Aasland, R, Finne, K, Veiseth, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The CW domain, a new histone recognition module in chromatin proteins.
Embo J., 30, 2011
|
|
2MLU
 
 | |
2MLV
 
 | |
2LXE
 
 | | S4wyild | | Descriptor: | Histone-lysine N-methyltransferase SUVR4 | | Authors: | Kristiansen, P, Rahman, M.A, Aalen, R.B. | | Deposit date: | 2012-08-20 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The arabidopsis histone methyltransferase SUVR4 binds ubiquitin via a domain with a four-helix bundle structure.
Biochemistry, 53, 2014
|
|
1G1J
 
 | |
1G1I
 
 | |
6FU8
 
 | |
2RLW
 
 | | Three-Dimensional Structure of the two Peptides that Constitute the Two-Peptide Bacteriocin Plantaracin EF | | Descriptor: | PlnF | | Authors: | Fimland, N, Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P. | | Deposit date: | 2007-08-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF
Biochim.Biophys.Acta, 1784, 2008
|
|
7N7X
 
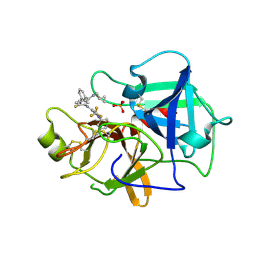 | | Crystal structure of BCX7353(ORLADEYO) in complex with human plasma kallikrein serine protease domain at 2.1 angstrom resolution | | Descriptor: | Orladeyo, PHOSPHATE ION, Plasma kallikrein light chain | | Authors: | Krishnan, R, Yarlagadda, B.S, Kotian, P, Polach, K.J, Zhang, W. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Berotralstat (BCX7353): Structure-Guided Design of a Potent, Selective, and Oral Plasma Kallikrein Inhibitor to Prevent Attacks of Hereditary Angioedema (HAE).
J.Med.Chem., 64, 2021
|
|
1B35
 
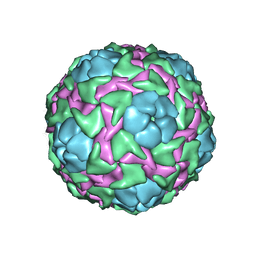 | | CRICKET PARALYSIS VIRUS (CRPV) | | Descriptor: | PROTEIN (CRICKET PARALYSIS VIRUS, VP1), VP2), ... | | Authors: | Tate, J.G, Liljas, L, Scotti, P.D, Christian, P.D, Lin, T.W, Johnson, J.E. | | Deposit date: | 1998-12-17 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cricket paralysis virus: the first view of a new virus family.
Nat.Struct.Biol., 6, 1999
|
|
2JUI
 
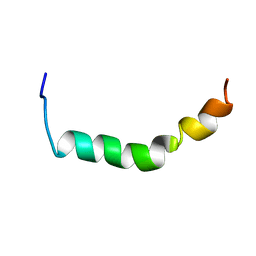 | | Three-Dimensional Structure of the two Peptides that Constitute the Two-Peptide Bacteriocin Plantaracin EF | | Descriptor: | PlnE | | Authors: | Fimland, N, Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P. | | Deposit date: | 2007-08-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF
Biochim.Biophys.Acta, 1784, 2008
|
|
2KEH
 
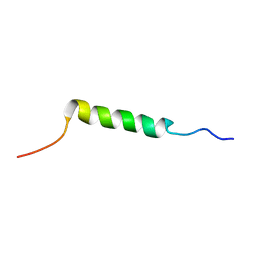 | |
2KEG
 
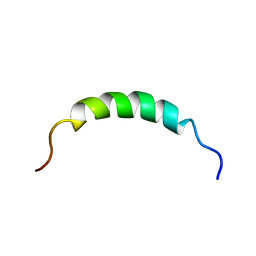 | | NMR structure of Plantaricin K in DPC-micelles | | Descriptor: | PlnK | | Authors: | Rogne, P, Haugen, M, Fimland, G, Nissen-Meyer, J, Kristiansen, P. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two-peptide bacteriocin plantaricin JK.
Peptides, 30, 2009
|
|
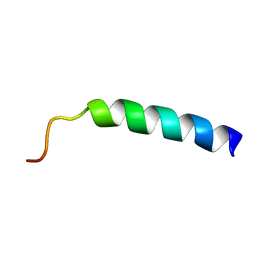
2KHG
 
 | |
8DEA
 
 | |
8DG6
 
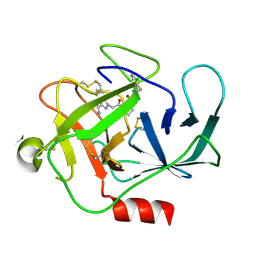 | |
8D95
 
 | |
