6ARC
 
 | |
6ARD
 
 | |
4TM6
 
 | |
4U6I
 
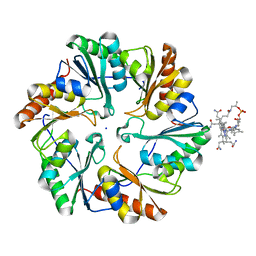 | | Crystal Structure of the EutL Microcompartment Shell Protein from Clostridium Perfringens Bound to Vitamin B12 | | Descriptor: | COBALAMIN, Ethanolamine utilization protein EutL, SODIUM ION | | Authors: | Thompson, M.C, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2014-07-29 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4LIW
 
 | |
4TME
 
 | |
4TLH
 
 | |
4OLO
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Clostridiales bacterium 1_7_47FAA | | Descriptor: | BMC domain protein | | Authors: | Thompson, M.C, Ahmed, H, McCarty, K.N, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
4EDI
 
 | | Disulfide bonded EutL from Clostridium perfringens | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
4FDZ
 
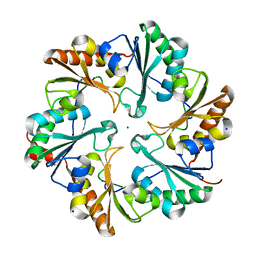 | | EutL from Clostridium perfringens, Crystallized Under Reducing Conditions | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
6W90
 
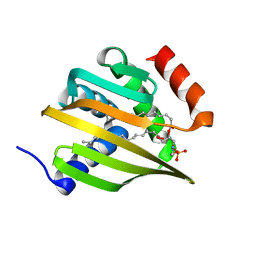 | | De novo designed NTF2 fold protein NT-9 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, NTF2 fold protein loop-helix-loop design NT-9 | | Authors: | Thompson, M.C, Pan, X, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2020-03-21 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expanding the space of protein geometries by computational design of de novo fold families.
Science, 369, 2020
|
|
5V5D
 
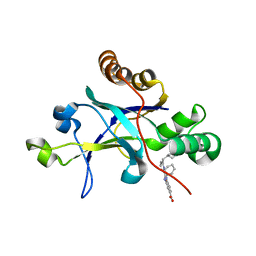 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 250) | | Descriptor: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-(phenylamino)benzoic acid, ORF 17 | | Authors: | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
5V5E
 
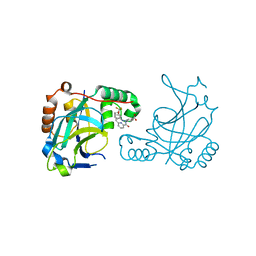 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 733) | | Descriptor: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-{[3-(trifluoromethoxy)phenyl]amino}benzoic acid, ORF 17 | | Authors: | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
4OLP
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Pectobacterium wasabiae | | Descriptor: | GrpU microcompartment shell protein | | Authors: | Wheatley, N.M, Thompson, M.C, Gidaniyan, S.D, Sawaya, M.R, Jorda, J, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
8VFW
 
 | | Crystal Structure of V113N D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG8
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFN
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 310K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG7
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG5
 
 | | Crystal Structure of V113N Variant of D-Dopachrome Tautomerase (D-DT) Bound with 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VDY
 
 | | Crystal Structure of Delta 114-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFL
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 290K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
9MYB
 
 | |
9MYA
 
 | |
4YUH
 
 | | Multiconformer synchrotron model of CypA at 150 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
