7ZMV
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMP
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-055 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-055, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMT
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMQ
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMS
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G4-043 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G4-043, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
4GG6
 
 | | Protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DQ alpha 1 chain, ... | | Authors: | Broughton, S.E, Theodossis, A, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease.
Immunity, 37, 2012
|
|
4GG8
 
 | | Immune Receptor | | Descriptor: | T-CELL RECEPTOR, SP3.4 ALPHA CHAIN, SP3.4 BETA CHAIN, ... | | Authors: | Broughton, S.E, Theodossis, A, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease.
Immunity, 37, 2012
|
|
2PTK
 
 | | CHICKEN SRC TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Williams, J.C, Wierenga, R. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions
J.Mol.Biol., 274, 1997
|
|
4Z7U
 
 | | S13 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DQ-alpha chain, MHC class II HLA-DQ-beta-1, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
4Z7W
 
 | | T316 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DQ8-glia-alpha1, MHC class II HLA-DQ-alpha chain, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
4Z7V
 
 | | L3-12 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DQ-alpha chain, MHC class II HLA-DQ-beta-1, ... | | Authors: | Petersen, J, Rossjohn, J, Reid, H.H, Koning, F. | | Deposit date: | 2015-04-08 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
J Immunol., 194, 2015
|
|
1B3G
 
 | |
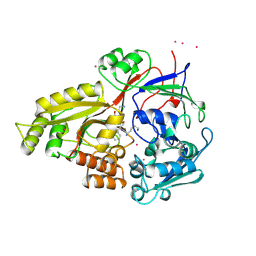
1B40
 
 | |
1B05
 
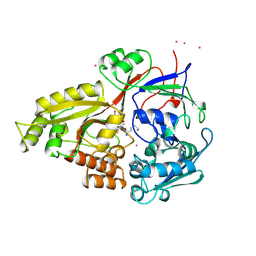 | |
1B4Z
 
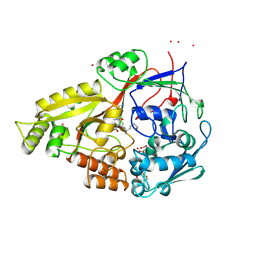 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KDK | | Descriptor: | ACETATE ION, PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), PROTEIN (PEPTIDE LYS-ASP-LYS), ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and calorimetric analysis of peptide binding to OppA protein.
J.Mol.Biol., 291, 1999
|
|
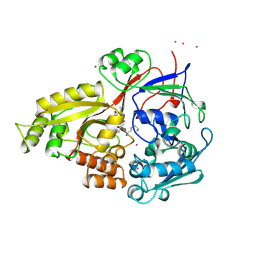
1B5J
 
 | |
1B51
 
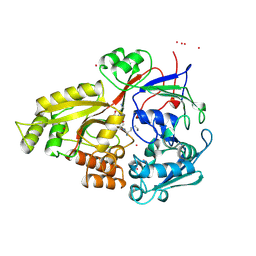 | |
2VX8
 
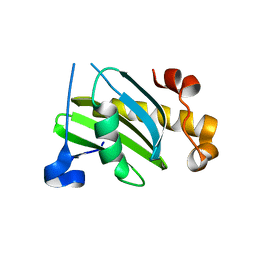 | | Vamp7 longin domain Hrb peptide complex | | Descriptor: | CHLORIDE ION, NUCLEOPORIN-LIKE PROTEIN RIP, VESICLE-ASSOCIATED MEMBRANE PROTEIN 7 | | Authors: | Evans, P.R, Owen, D.J, Luzio, J.P. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Sorting of the Snare Vamp7 Into Endocytic Clathrin-Coated Vesicles by the Arfgap Hrb.
Cell(Cambridge,Mass.), 134, 2008
|
|
2XFB
 
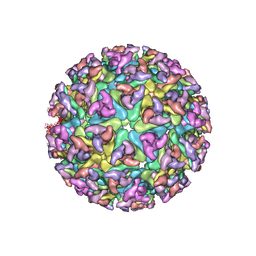 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SINDBIS VIRUS cryo- EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
2XFC
 
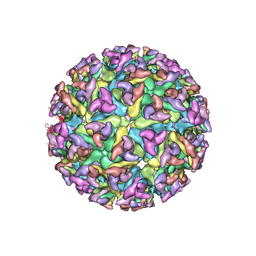 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SEMLIKI FOREST VIRUS cryo-EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
7GB5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDG-MED-0da5ad92-2 (Mpro-x10201) | | Descriptor: | 2-(3-chlorophenyl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-6 (Mpro-x10172) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-3c65e9ce-2 (Mpro-x12719) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(4-cyclopropylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-49816e9b-2 (Mpro-x10334) | | Descriptor: | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-ce760d3f-4 (Mpro-x12723) | | Descriptor: | (4R)-6-chloro-N-(2-oxo-2lambda~5~-isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
