4UFC
 
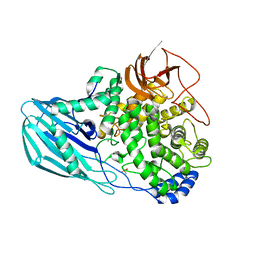 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
5G2U
 
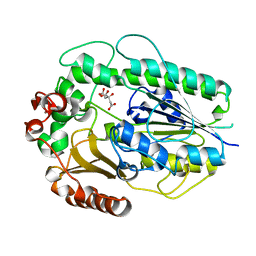 | | Structure of BT1596,a 2-O GAG sulfatase | | Descriptor: | 2-O GLYCOSAMINOGLYCAN SULFATASE, CITRIC ACID, ZINC ION | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2T
 
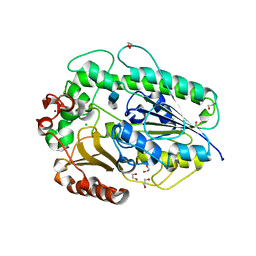 | | BT1596 in complex with its substrate 4,5 unsaturated uronic acid alpha 1,4 D-Glucosamine-2-N, 6-O-disulfate | | Descriptor: | 1,2-ETHANEDIOL, 2-O GLYCOSAMINOGLYCAN SULFATASE, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2V
 
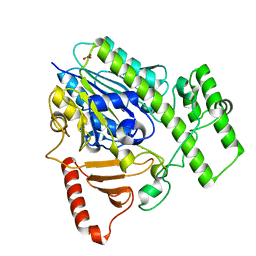 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6S20
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33336S-sulf) | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, N-acetylgalactosamine-6-O-sulfatase, ... | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
6S21
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
5NOA
 
 | | Polysaccharide Lyase BACCELL_00875 | | Descriptor: | Family 88 glycosyl hydrolase | | Authors: | Cartmell, A, Munoz-Munoz, J, Terrapon, N, Basle, A, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins.
J. Biol. Chem., 292, 2017
|
|
5MUM
 
 | | Glycoside Hydrolase BACINT_00347 | | Descriptor: | 1,2-ETHANEDIOL, BACINT_00347, PENTAETHYLENE GLYCOL | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MUK
 
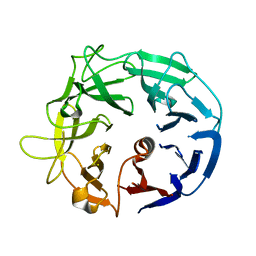 | | Glycoside Hydrolase BT3686 | | Descriptor: | Neuraminidase | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MVH
 
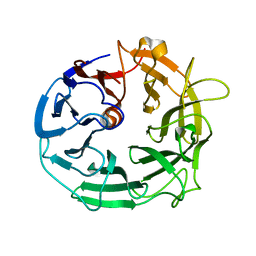 | | Glycoside Hydrolase BACCELL_00856 | | Descriptor: | BACCELL_00856 | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MUL
 
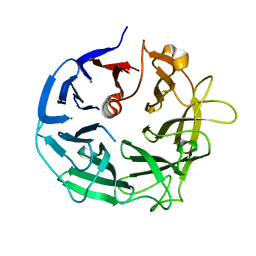 | | Glycoside Hydrolase BT3686 bound to Glucuronic Acid | | Descriptor: | Neuraminidase, beta-D-glucopyranuronic acid | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NO8
 
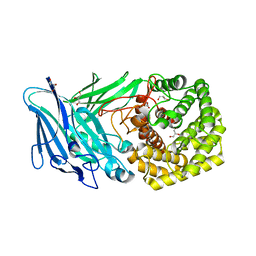 | | Polysaccharide Lyase BACCELL_00875 | | Descriptor: | BACCELL_00875, GLYCEROL | | Authors: | Cartmell, A, Munoz-Munoz, J, Terrapon, N, Basle, A, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-28 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins.
J. Biol. Chem., 292, 2017
|
|
5NOK
 
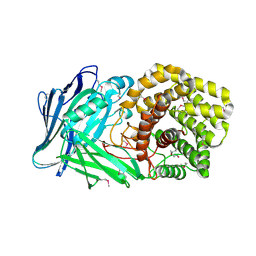 | | Polysaccharide Lyase BACCELL_00875 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BACCELL_00875 | | Authors: | Cartmell, A, Munoz-Munoz, J, Terrapon, N, Basle, A, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins.
J. Biol. Chem., 292, 2017
|
|
6XYC
 
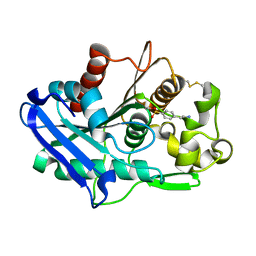 | | Truncated form of carbohydrate esterase from gut microbiota | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Acetyl xylan esterase | | Authors: | Penttinen, L, Hakulinen, N, Master, E. | | Deposit date: | 2020-01-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
6TKX
 
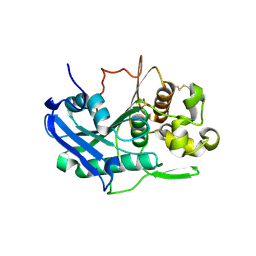 | | Carbohydrate esterase from gut microbiota | | Descriptor: | Carbohydrate esterase, SULFATE ION | | Authors: | Penttinen, L, Hakulinen, N, Master, R.E. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polysaccharide utilization loci-driven enzyme discovery reveals BD-FAE: a bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans.
Biotechnol Biofuels, 14, 2021
|
|
7JVI
 
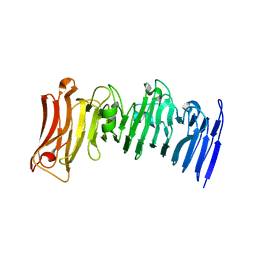 | | Crystal structure of a beta-helix domain retrieved from capybara gut metagenome | | Descriptor: | Beta-helix domain, CALCIUM ION | | Authors: | Martins, M.P, Genoroso, W.C, Domingues, M.N, Persinoti, G.F, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2020-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gut microbiome of the largest living rodent harbors unprecedented enzymatic systems to degrade plant polysaccharides.
Nat Commun, 13, 2022
|
|
8D89
 
 | | Crystal structure of a novel GH5 enzyme retrieved from capybara gut metagenome | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, M.P, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glycoside hydrolase subfamily GH5_57 features a highly redesigned catalytic interface to process complex hetero-beta-mannans.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8DGE
 
 | | BoGH13ASus from Bacteroides ovatus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Alpha amylase, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
8DL1
 
 | | BoGH13ASus-E523Q from Bacteroides ovatus bound to maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha amylase, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
8DL2
 
 | | BoGH13ASus from Bacteroides ovatus bound to acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
5OPJ
 
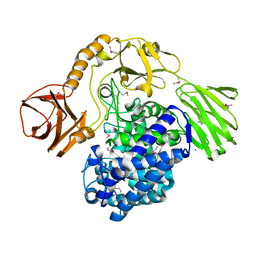 | | Beta-L-arabinofuranosidase | | Descriptor: | Rhamnogalacturonan lyase, ZINC ION, alpha-L-arabinofuranose | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-08-10 | | Release date: | 2018-02-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
4AK2
 
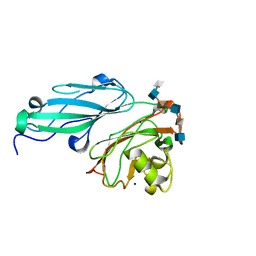 | | Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BT_4661, SODIUM ION | | Authors: | Lowe, E.C, Basle, A, Czjzek, M, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2012-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4AK1
 
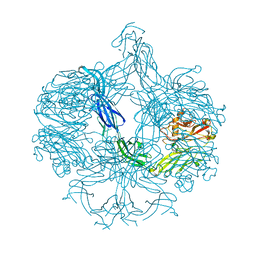 | | Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus | | Descriptor: | BT_4661, SODIUM ION | | Authors: | Lowe, E.C, Basle, A, Czjzek, M, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2012-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6EON
 
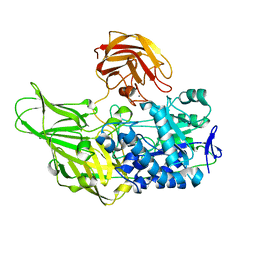 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUI
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
