8J4G
 
 | | Crystal structure of 11JD mutant-I62N | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of duck MHC 11JD mutant-I62N
To Be Published
|
|
8JQT
 
 | | Crystal structure of U03-GPAKGIEYD | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of duck MHC U03
To Be Published
|
|
8KB0
 
 | | Crystal structure of 01JD-AEAIIVAMV | | Descriptor: | Beta2-microglobulin, MHC class I antigen alpha chain, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KB1
 
 | | Crystal structure of 11JD | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
8KCV
 
 | | Crystal structure of UDA01-CAAMDDFQL | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of duck MHC UDA01
To Be Published
|
|
8WO9
 
 | |
1S2H
 
 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1KLQ
 
 | | The Mad2 Spindle Checkpoint Protein Undergoes Similar Major Conformational Changes upon Binding to Either Mad1 or Cdc20 | | Descriptor: | MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A, Mad2-binding peptide | | Authors: | Luo, X, Tang, Z, Rizo, J, Yu, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20.
Mol.Cell, 9, 2002
|
|
8T5K
 
 | | Crystal structure of STING CTD in complex with BDW-OH | | Descriptor: | Stimulator of interferon genes protein, {[(4S)-8,9-dimethylthieno[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-3-yl]sulfanyl}acetic acid | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8T5L
 
 | | Crystal structure of STING CTD in complex with 2'3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
4R5Y
 
 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | Descriptor: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | Authors: | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | Deposit date: | 2014-08-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
8HVS
 
 | |
9KSH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 8 | | Descriptor: | 3-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-(2-methoxyethoxy)pyridin-3-yl]-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-4-methyl-benzenecarbonitrile, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
4DZO
 
 | |
9KR5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 3 | | Descriptor: | (6~{E})-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-[(3~{S})-oxolan-3-yl]oxypyridin-3-yl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSI
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with compound 5 | | Descriptor: | (6E)-1-[[5-chloranyl-4-fluoranyl-2-(4-fluoranylphenoxy)phenyl]methyl]-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methoxypyridin-3-yl)-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 10 | | Descriptor: | 3C-like proteinase, 4-[4-chloranyl-2-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methylpyridin-3-yl)-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-5-fluoranyl-phenoxy]-2-fluoranyl-benzenecarbonitrile | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
2QYF
 
 | |
6SMB
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | Tyrosine-protein kinase JAK1, ~{N}-[3-[2-[(3-methoxy-1-methyl-pyrazol-4-yl)amino]-5-methyl-pyrimidin-4-yl]-1~{H}-indol-7-yl]-2-methyl-pyridine-3-carboxamide | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6SM8
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-6-[(3~{S})-3-[(1~{S})-2-cyano-1-[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]ethyl]pyrrolidin-1-yl]benzenecarbonitrile, Tyrosine-protein kinase JAK1 | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
4JOL
 
 | |
6LAT
 
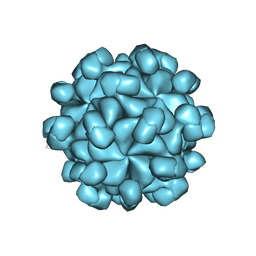 | | The cryo-EM structure of HEV VLP | | Descriptor: | Protein ORF2 | | Authors: | Zheng, Q, He, M, Li, S. | | Deposit date: | 2019-11-13 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Viral neutralization by antibody-imposed physical disruption.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6LB0
 
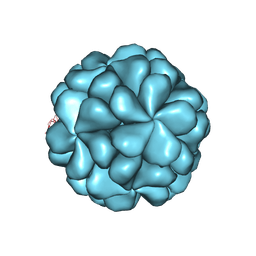 | |
5ZBZ
 
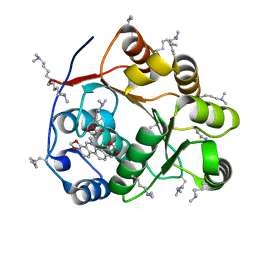 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
