7BSX
 
 | | SDR protein NapW-NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-03-31 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
7BTM
 
 | | SDR protein/resistance protein NapW | | Descriptor: | Short chain dehydrogenase | | Authors: | Wen, W.H, Tang, G.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08311653 Å) | | Cite: | Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics.
Nat Commun, 12, 2021
|
|
7LXJ
 
 | |
7LXH
 
 | | Bacillus cereus DNA glycosylase AlkD bound to a CC1065-adenine nucleobase adduct and DNA containing an abasic site | | Descriptor: | 7-{7-[(1R)-1-{[(4P)-6-amino-3H-purin-3-yl]methyl}-5-hydroxy-8-methyl-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carbonyl]-4-hydroxy-5-methoxy-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carbonyl}-4-hydroxy-5-methoxy-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carboxamide, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*AP*(ORP)P*GP*GP*C)-3'), ... | | Authors: | Mullins, E.A, Eichman, B.F. | | Deposit date: | 2021-03-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Structural evolution of a DNA repair self-resistance mechanism targeting genotoxic secondary metabolites.
Nat Commun, 12, 2021
|
|
7XH2
 
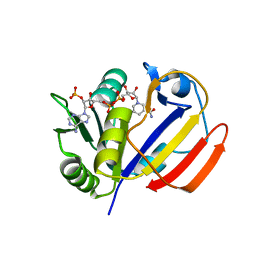 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
7XH4
 
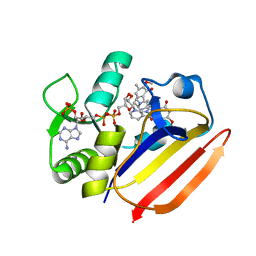 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis complex with safracin A | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Safracin A, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Zhang, Y, Yang, D, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
8GP7
 
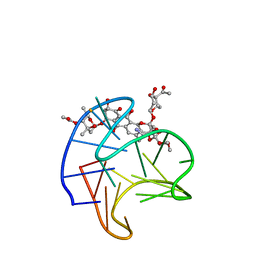 | | Structure of Trioxacarcin A covalently bound to RET G4-DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), Trioxacarcin A, bound form | | Authors: | Yin, S, Cao, C. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Trioxacarcin A Interactions with G-Quadruplex DNA Reveal Its Potential New Targets as an Anticancer Agent.
J.Med.Chem., 66, 2023
|
|
8HIV
 
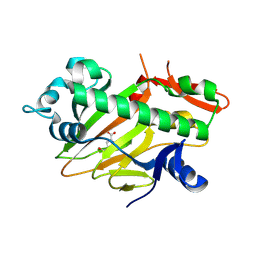 | | The structure of apo-SoBcmB with Fe(II) and AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Fe/2OG dependent dioxygenase | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2022-11-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2000308 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
5Z4F
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with SUM | | Descriptor: | 1,8-dihydroxy-2-[(4R)-4-hydroxy-4-methyl-3-oxohexanoyl]-3-methylanthracene-9,10-dione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z4E
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 | | Descriptor: | TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YSW
 
 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5YSM
 
 | | Crystal Structure Analysis of Rif16 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5X5M
 
 | |
5Z36
 
 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with PDM | | Descriptor: | 11-hydroxy-2-[(2S)-2-hydroxybutan-2-yl]-5-methyl-4H-anthra[1,2-b]pyran-4,7,12-trione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7V3E
 
 | | The complex structure of soBcmB and its intermediate product 1a | | Descriptor: | (3S,6Z)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2U
 
 | | The complex structure of SoBcmB and its product 2f | | Descriptor: | (1S,5S,6S)-5-methyl-1-[(1S,2S)-2-methyl-1,2,3-tris(oxidanyl)propyl]-2-oxa-7,9-diazabicyclo[4.2.2]decane-8,10-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.00009918 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V34
 
 | | The complex structure of soBcmB and its product 1d | | Descriptor: | (3S,4S,5S,8S)-8-[(2S)-butan-2-yl]-3-methyl-3,4-bis(oxidanyl)-1-oxa-7,10-diazaspiro[4.5]decane-6,9-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.000169 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V36
 
 | | The complex structure of soBcmB and its intermediate product 2a | | Descriptor: | (3Z,6S)-3-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]-6-[(2S)-4-oxidanylbutan-2-yl]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.96009171 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3N
 
 | | The complex structure of soBcmB-D307A and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.850011 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2X
 
 | | The complex structure of soBcmB and its substrate 1 | | Descriptor: | (3S,6S)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propyl]piperazine-2,5-dion, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.08387423 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3O
 
 | | The structure of Se-SoBcmB with Fe(II)and AKG | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83004665 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
