5WED
 
 | |
7YPD
 
 | |
4GA4
 
 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | Descriptor: | PHOSPHATE ION, Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4GA5
 
 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | Descriptor: | Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
3VLU
 
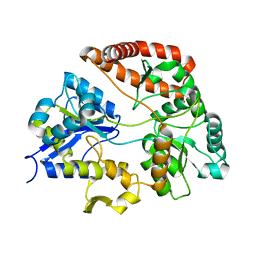 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with saturated trimannuronate | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VU4
 
 | |
3VVP
 
 | | Crystal structure of MATE in complex with Br-NRF | | Descriptor: | 6-bromo-1-ethyl-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVR
 
 | | Crystal structure of MATE in complex with MaD5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
6LEP
 
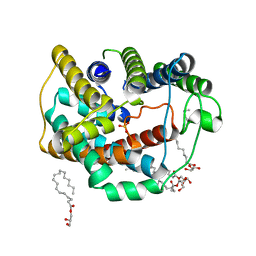 | | Crystal structure of thiosulfate transporter YeeE inactive mutant - C91A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Sugano, Y, Takeuchi, A, Uchino, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
6LEO
 
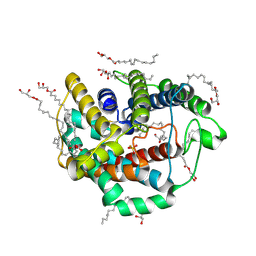 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
8HP3
 
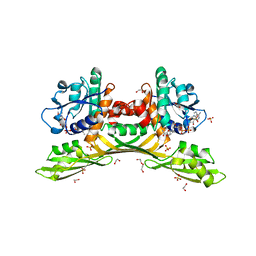 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HP0
 
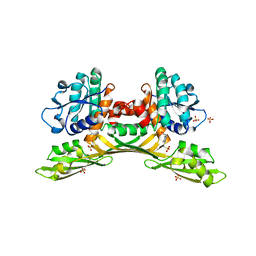 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
7DQK
 
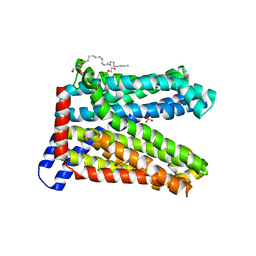 | | A nicotine MATE transporter, Nicotiana tabacum MATE2 (NtMATE2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, Y, Tsukazaki, T, Sasaki, A, Iwaki, S. | | Deposit date: | 2020-12-24 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a nicotine MATE transporter provide insight into its mechanism of substrate transport.
Febs Lett., 595, 2021
|
|
7X5H
 
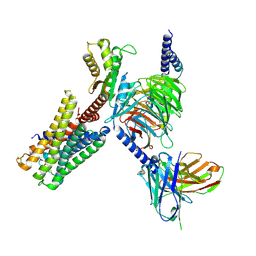 | | Serotonin 5A (5-HT5A) receptor-Gi protein complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tan, Y, Xu, P, Huang, S, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-03-04 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the ligand binding and G i coupling of serotonin receptor 5-HT 5A .
Cell Discov, 8, 2022
|
|
5ZOI
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase | | Descriptor: | Alpha-(1,3)-fucosyltransferase FucT, [[(2S,3R,4S,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5R,6R)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Tan, Y, Yang, G. | | Deposit date: | 2018-04-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Directed evolution of an alpha 1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method.
Sci Adv, 5, 2019
|
|
1WU7
 
 | | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Tanaka, Y, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum
To be Published
|
|
1WV3
 
 | |
2DG0
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | DrP35 | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DG1
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus, complexed with Ca2+ | | Descriptor: | CALCIUM ION, DrP35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DSO
 
 | | Crystal structure of D138N mutant of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Drp35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-07-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
1J08
 
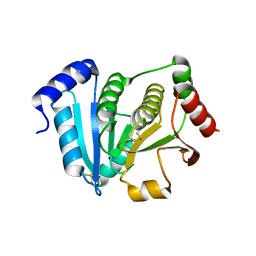 | | Crystal structure of glutaredoxin-like protein from Pyrococcus horikoshii | | Descriptor: | glutaredoxin-like protein | | Authors: | Tanaka, Y, Tanabe, E, Tsumoto, K, Kumagai, I, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-11 | | Release date: | 2003-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein disulfide isomerase from hyperthermophile as an additives of refolding of an immunoglobulin-folded protein
To be Published
|
|
1FVM
 
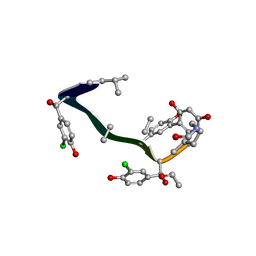 | | Complex of vancomycin with DI-acetyl-LYS-D-ALA-D-ALA | | Descriptor: | DI-ACETYL-LYS-D-ALA-D-ALA, VANCOMYCIN, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Nitanai, Y, Kakoi, K, Aoki, K. | | Deposit date: | 2000-09-20 | | Release date: | 2000-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Complexes between Vancomycin and Cell-Wall Precursor Analogs.
J.Mol.Biol., 385, 2009
|
|
1HLV
 
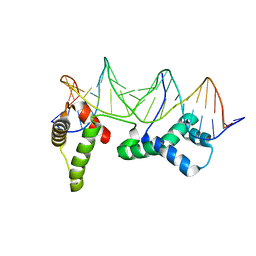 | | CRYSTAL STRUCTURE OF CENP-B(1-129) COMPLEXED WITH THE CENP-B BOX DNA | | Descriptor: | CENP-B BOX DNA, MAJOR CENTROMERE AUTOANTIGEN B | | Authors: | Tanaka, Y, Nureki, O, Kurumizaka, H, Fukai, S, Kawaguchi, S, Ikuta, M, Iwahara, J, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-04 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA.
EMBO J., 20, 2001
|
|
2DGD
 
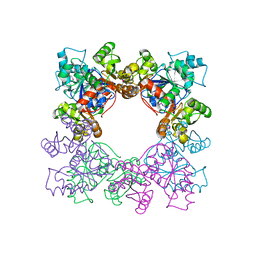 | | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii | | Descriptor: | 223aa long hypothetical arylmalonate decarboxylase, GLYCEROL | | Authors: | Tanaka, Y, Sasaki, T, Tanabe, E, Yao, M, Tanaka, I, Kumagai, I, Tsumoto, K. | | Deposit date: | 2006-03-10 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii
To be Published
|
|
