3ERM
 
 | |
2QLT
 
 | | Crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p, from Saccharomyces cerevisiae | | Descriptor: | (DL)-glycerol-3-phosphatase 1, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p from Saccharomyces cerevisiae.
To be Published
|
|
3FC7
 
 | |
2QZI
 
 | | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311. | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a conserved protein of unknown function from Streptococcus thermophilus LMG 18311.
To be Published
|
|
2QMX
 
 | | The crystal structure of L-Phe inhibited prephenate dehydratase from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHENYLALANINE, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2QIP
 
 | | Crystal structure of a protein of unknown function VPA0982 from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, Protein of unknown function VPA0982 | | Authors: | Tan, K, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of a protein of unknown function, VPA0982 from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
3FG9
 
 | | The crystal structure of an universal stress protein UspA family protein from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Tan, K, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of an universal stress protein UspA family protein from Lactobacillus plantarum WCFS1
To be Published
|
|
3FK8
 
 | |
3F1B
 
 | | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TetR-like transcriptional regulator | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3FFH
 
 | |
2QZ7
 
 | | The crystal structure of a homologue of telluride resistance protein (TerD), SCO6318 from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein SCO6318 | | Authors: | Tan, K, Xu, X, Zheng, Z, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a homologue of telluride resistance protein (TerD), SCO6318 from Streptomyces coelicolor A3(2).
To be Published
|
|
2QL3
 
 | | Crystal structure of the C-terminal domain of a probable LysR family transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | PHOSPHATE ION, Probable transcriptional regulator, LysR family protein | | Authors: | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-12 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of the C-terminal domain of a probable LysR family transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3F5B
 
 | | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside N(6')acetyltransferase, ... | | Authors: | Tan, K, Wu, R, Perez, V, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of aminoglycoside N(6')acetyltransferase from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
3F5R
 
 | | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p). | | Descriptor: | CHLORIDE ION, FACT complex subunit POB3, FORMIC ACID, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a subunit of the heterodimeric FACT complex (Spt16p-Pob3p).
To be Published
|
|
4Q6T
 
 | | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5 | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tan, K, Mack, J.C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5
To be Published
|
|
4PYS
 
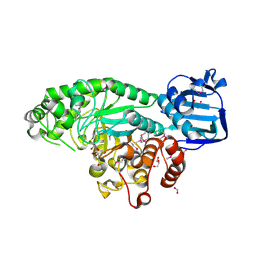 | | The crystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343 | | Descriptor: | FORMIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | The crystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343
To be Published
|
|
2R8W
 
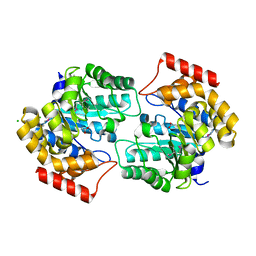 | | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETATE ION, AGR_C_1641p, CHLORIDE ION | | Authors: | Tan, K, Dong, A, Xu, X, Gu, J, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of dihydrodipicolinate synthase (Atu0899) from Agrobacterium tumefaciens str. C58.
To be Published
|
|
4Q7Q
 
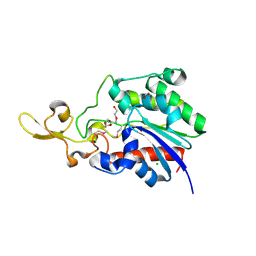 | | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588 | | Descriptor: | CHLORIDE ION, FORMIC ACID, Lipolytic protein G-D-S-L family, ... | | Authors: | Tan, K, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588
To be Published
|
|
2R5R
 
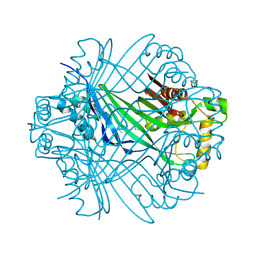 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
3FBY
 
 | |
2R8B
 
 | | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | SULFATE ION, Uncharacterized protein Atu2452 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-10 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
2R5S
 
 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
6BB9
 
 | | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-4-deoxychorismate lyase, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-17 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2
To Be Published
|
|
