6OR6
 
 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (tail region) | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6ORB
 
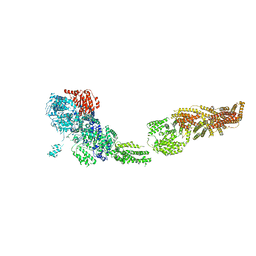 | | Full-length S. pombe Mdn1 in the presence of ATP and Rbin-1 | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6OR5
 
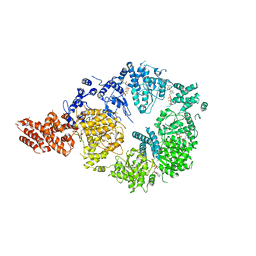 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (ring region) | | Descriptor: | Midasin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
1NX4
 
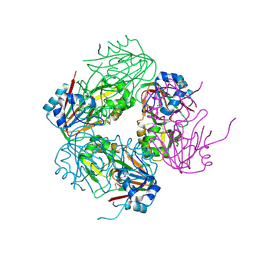 | | The crystal structure of carbapenem synthase (CarC) | | Descriptor: | 2-OXOGLUTARIC ACID, Carbapenem synthase, FE (III) ION | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-08 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
1NX8
 
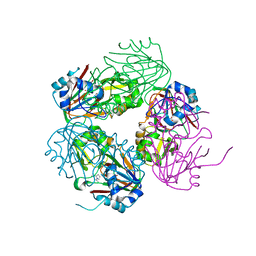 | | Structure of carbapenem synthase (CarC) complexed with N-acetyl proline | | Descriptor: | 1-ACETYL-L-PROLINE, 2-OXOGLUTARIC ACID, Carbapenem synthase, ... | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
2NQO
 
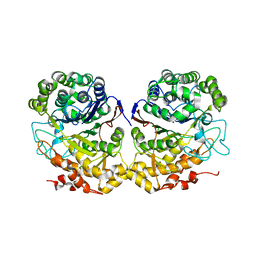 | | Crystal Structure of Helicobacter pylori gamma-Glutamyltranspeptidase | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Boanca, G, Sand, A, Okada, T, Suzuki, H, Kumagai, H, Fukuyama, K, Barycki, J.J. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad.
J.Biol.Chem., 282, 2007
|
|
8IHJ
 
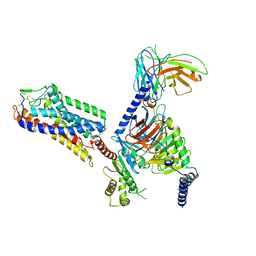 | | Cryo-EM structure of HCA3-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHF
 
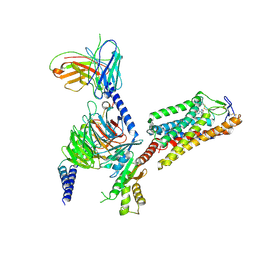 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
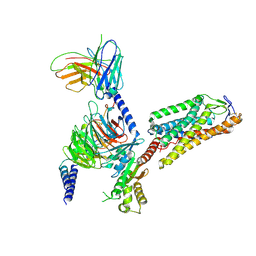 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHI
 
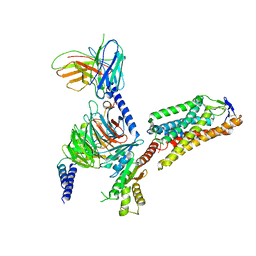 | | Cryo-EM structure of HCA2-Gi complex with acifran | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | Descriptor: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
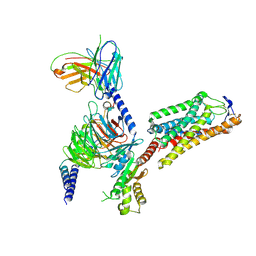 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
3AAK
 
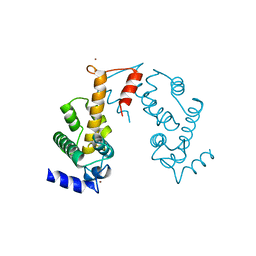 | | Crystal structure of Zn2+-bound form of des3-20ALG-2F122A | | Descriptor: | Programmed cell death protein 6, ZINC ION | | Authors: | Inuzuka, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
5DPT
 
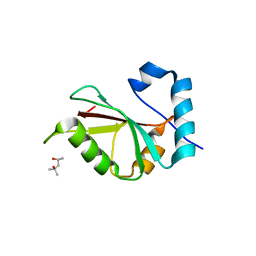 | | Crystal structure of PLEKHM1 LIR-fused human GABARAPL1_2-117 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Pleckstrin homology domain-containing family M member 1, Gamma-aminobutyric acid receptor-associated protein-like 1,Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Ravichandran, A.C, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional analysis of the GABARAP interaction motif (GIM).
EMBO Rep., 18, 2017
|
|
5DPS
 
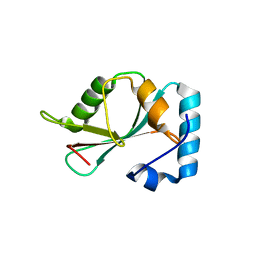 | |
5DPW
 
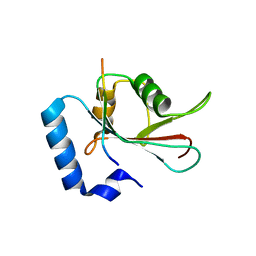 | |
5DPR
 
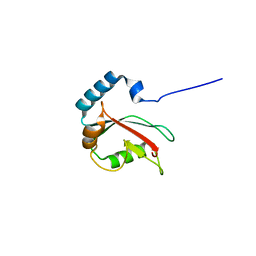 | |
2YR4
 
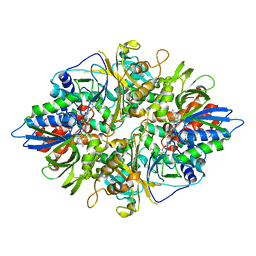 | | Crystal structure of L-phenylalanine oxiase from Psuedomonas sp. P-501 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pro-enzyme of L-phenylalanine oxidase, SULFATE ION | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2YR6
 
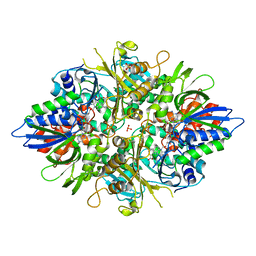 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2YR5
 
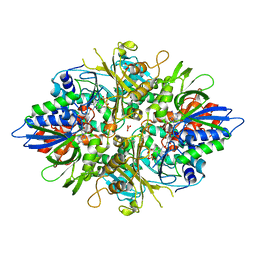 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Pro-enzyme of L-phenylalanine oxidase, ... | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Hikima, T, Yamamoto, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2ZPI
 
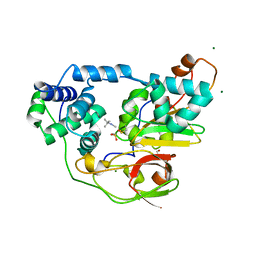 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 440min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPB
 
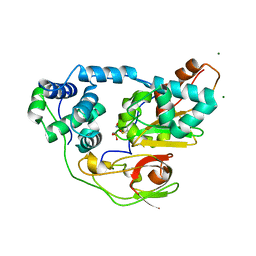 | | nitrosylated Fe-type nitrile hydratase | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-09 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPH
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 340min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPG
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 120min at 293K | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
