8X38
 
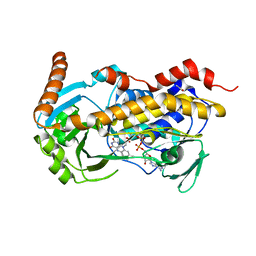 | | Crystal Structure of Decarboxylative Vanillate 1-Hydroxylase from Phanerochaete chrysosporium | | Descriptor: | ACETATE ION, Decarboxylative Vanillate 1-Hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Suzuki, H, Mori, R, Ishida, T, Igarashi, K, Shimizu, M. | | Deposit date: | 2023-11-12 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decarboxylative Vanillate 1-Hydroxylase from Phanerochaete chrysosporium
To Be Published
|
|
3AAJ
 
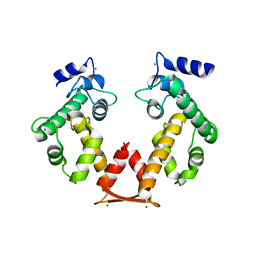 | | Crystal structure of Ca2+-bound form of des3-23ALG-2deltaGF122 | | Descriptor: | CALCIUM ION, Programmed cell death protein 6 | | Authors: | Suzuki, H, Inuzuka, T, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
8HUJ
 
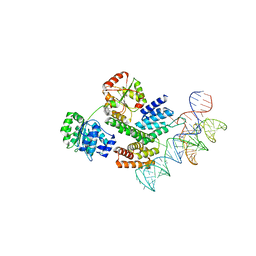 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), ... | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
8J7R
 
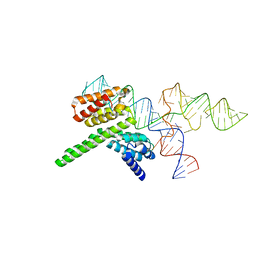 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused) | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), MAGNESIUM ION | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
3AD8
 
 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 in complex with pyrrole 2-carboxylate | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3ADA
 
 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 in complex with sulfite | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3AD7
 
 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 in complex with methylthio acetate | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3AD9
 
 | | Heterotetrameric Sarcosine Oxidase from Corynebacterium sp. U-96 sarcosine-reduced form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, H, Moriguchi, T, Ida, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Channeling and conformational changes in the heterotetrameric sarcosine oxidase from Corynebacterium sp. U-96.
J.Biochem., 148, 2010
|
|
3AYL
 
 | |
3VTW
 
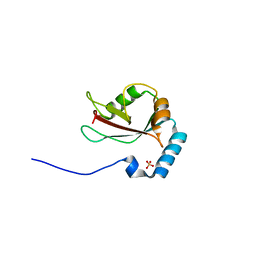 | | Crystal structure of T7-tagged Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VTU
 
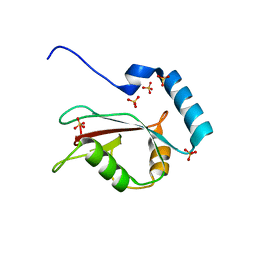 | | Crystal structure of human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VTV
 
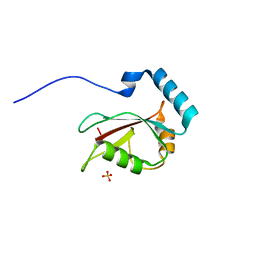 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3X0W
 
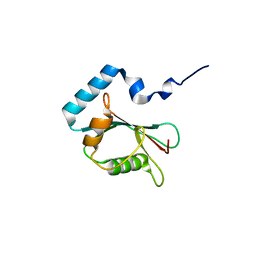 | | Crystal structure of PLEKHM1 LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, McEwan, D.G, Popovic, D, Gubas, A, Terawaki, S, Stadel, D, Coxon, F, Stegmann, D.M, Bhogaraju, S, Maddi, K, Kirchhoff, A, Gatti, E, Helfrich, M.H, Behrends, C, Pierre, P, Dikic, I, Wakatsuki, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
Mol.Cell, 57, 2015
|
|
1C7G
 
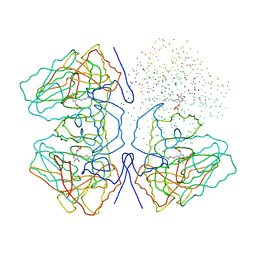 | |
3IYZ
 
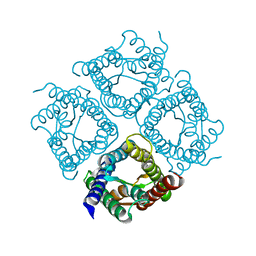 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
4TLJ
 
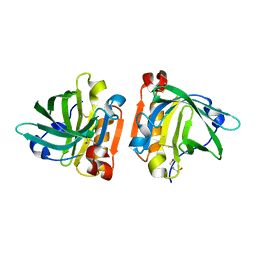 | | Ultra-high resolution crystal structure of caprine Beta-lactoglobulin | | Descriptor: | 1,4-BUTANEDIOL, Beta-lactoglobulin | | Authors: | Crowther, J.M, Jameson, G.B, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ultra-high resolution crystal structure of recombinant caprine beta-lactoglobulin.
Febs Lett., 588, 2014
|
|
6UT6
 
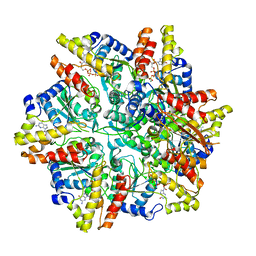 | | Cryo-EM structure of the Escherichia coli McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT4
 
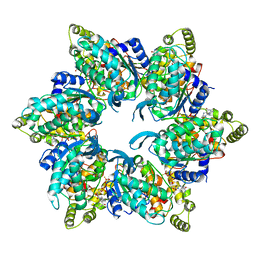 | | Cryo-EM structure of the asymmetric AAA+ domain hexamer from Thermococcus gammatolerans McrB | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, MAGNESIUM ION | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT7
 
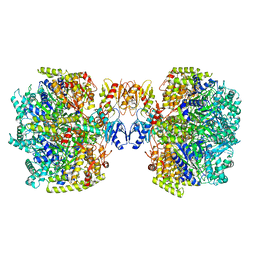 | | Fitted model for the tetradecameric assembly of Thermococcus gammatolerans McrB AAA+ hexamers with bound McrC | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT5
 
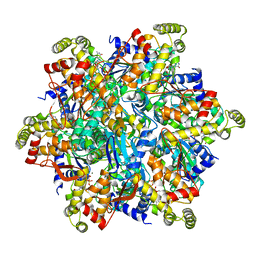 | | Cryo-EM structure of the Thermococcus gammatolerans McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UZL
 
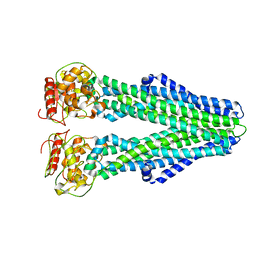 | | Cryo-EM structure of nucleotide-free MsbA reconstituted into peptidiscs, conformation 2 | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Angiulli, G, Walz, T, Dhupar, H.S, Suzuki, H, Wason, I.S, Duong Van Hoa, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins.
Elife, 9, 2020
|
|
6UZH
 
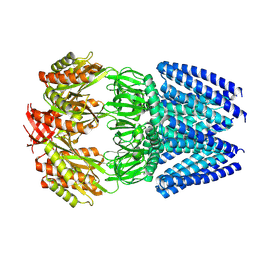 | | Cryo-EM structure of mechanosensitive channel MscS reconstituted into peptidiscs | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Angiulli, G, Walz, T, Dhupar, H.S, Suzuki, H, Wason, I.S, Duong Van Hoa, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins.
Elife, 9, 2020
|
|
6UZ2
 
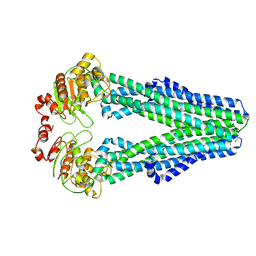 | | Cryo-EM structure of nucleotide-free MsbA reconstituted into peptidiscs, conformation 1 | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Angiulli, G, Walz, T, Dhupar, H.S, Suzuki, H, Wason, I.S, Duong Van Hoa, F. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins.
Elife, 9, 2020
|
|
6UT8
 
 | | Refined half-complex from tetradecameric assembly of Thermococcus gammatolerans McrB AAA+ hexamers with bound McrC | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase subunit of restriction endonuclease, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
8GCL
 
 | | Cryo-EM structure of hAQP2 in DDM | | Descriptor: | Aquaporin-2 | | Authors: | Kamegawa, A, Suzuki, S, Nishikawa, K, Numoto, N, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2023-03-02 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural analysis of the water channel AQP2 by single-particle cryo-EM.
J.Struct.Biol., 215, 2023
|
|
