1Q3V
 
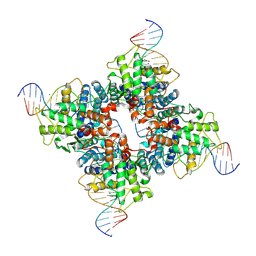 | | Crystal structure of a wild-type Cre recombinase-loxP synapse: phosphotyrosine covalent intermediate | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-08-01 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
1QPH
 
 | |
1EN7
 
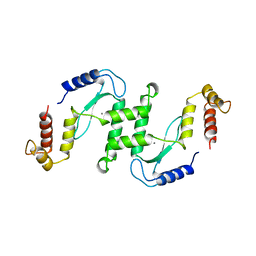 | | ENDONUCLEASE VII (ENDOVII) FROM PHAGE T4 | | Descriptor: | CALCIUM ION, RECOMBINATION ENDONUCLEASE VII, ZINC ION | | Authors: | Raaijmakers, H, Vix, O, Toro, I, Suck, D. | | Deposit date: | 1999-02-07 | | Release date: | 2000-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture.
EMBO J., 18, 1999
|
|
2HRK
 
 | |
2HSN
 
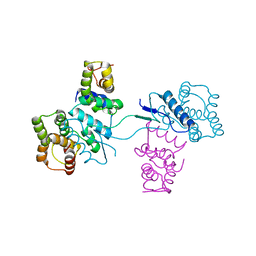 | | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes | | Descriptor: | GU4 nucleic-binding protein 1, Methionyl-tRNA synthetase, cytoplasmic | | Authors: | Simader, H, Koehler, C, Basquin, J, Suck, D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes.
Nucleic Acids Res., 34, 2006
|
|
2HSM
 
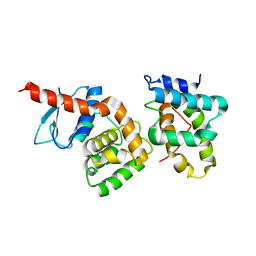 | |
1H64
 
 | |
1HLO
 
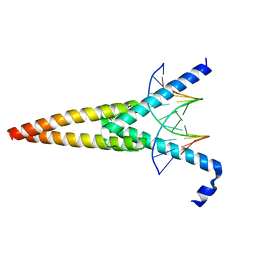 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1I4K
 
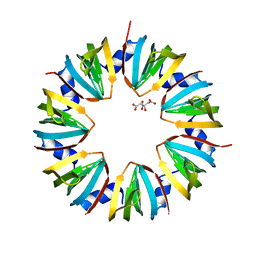 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1HK9
 
 | |
1LFB
 
 | | THE X-RAY STRUCTURE OF AN ATYPICAL HOMEODOMAIN PRESENT IN THE RAT LIVER TRANSCRIPTION FACTOR LFB1(SLASH)HNF1 AND IMPLICATIONS FOR DNA BINDING | | Descriptor: | LIVER TRANSCRIPTION FACTOR (LFB1) | | Authors: | Ceska, T.A, Lamers, M, Monaci, P, Nicosia, A, Cortese, R, Suck, D. | | Deposit date: | 1993-06-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding.
EMBO J., 12, 1993
|
|
1M8V
 
 | | Structure of Pyrococcus abyssii Sm Protein in Complex with a Uridine Heptamer | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', CALCIUM ION, PUTATIVE SNRNP SM-LIKE PROTEIN, ... | | Authors: | Thore, S, Mayer, C, Sauter, C, Weeks, S, Suck, D. | | Deposit date: | 2002-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Pyrococcus abyssii Sm core and its Complex with RNA: Common Features of RNA-binding in Archaea and Eukarya
J.Biol.Chem., 278, 2003
|
|
1XO1
 
 | | T5 5'-EXONUCLEASE MUTANT K83A | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Suck, D, Sayers, J.R. | | Deposit date: | 1998-11-19 | | Release date: | 1999-04-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis of conserved lysine residues in bacteriophage T5 5'-3' exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1UT5
 
 | |
1UT8
 
 | |
1TH7
 
 | |
1UOC
 
 | | X-ray structure of the RNase domain of the yeast Pop2 protein | | Descriptor: | CALCIUM ION, POP2, XENON | | Authors: | Thore, S, Mauxion, F, Seraphin, B, Suck, D. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure and Activity of the Yeast Pop2 Protein: A Nuclease Subunit of the Mrna Deadenylase Complex
Embo Rep., 4, 2003
|
|
4SBV
 
 | |
3PRK
 
 | | INHIBITION OF PROTEINASE K BY METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE. AN X-RAY STUDY AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE, PROTEINASE K | | Authors: | Wolf, W.M, Bajorath, J, Mueller, A, Raghunathan, S, Singh, T.P, Hinrichs, W, Saenger, W. | | Deposit date: | 1991-08-07 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of proteinase K by methoxysuccinyl-Ala-Ala-Pro-Ala-chloromethyl ketone. An x-ray study at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
3GOX
 
 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I in the absence of EDTA | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', PENTAETHYLENE GLYCOL, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
2PRK
 
 | |
4WZJ
 
 | | Spliceosomal U4 snRNP core domain | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Leung, A.K.W, Nagai, K, Li, J. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis.
Nature, 473, 2011
|
|
3FC3
 
 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
1YVN
 
 | | THE YEAST ACTIN VAL 159 ASN MUTANT COMPLEX WITH HUMAN GELSOLIN SEGMENT 1. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Vorobiev, S.M, Belmont, L.D, Drubin, D.G, Almo, S.C. | | Deposit date: | 1999-03-23 | | Release date: | 2000-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of yeast actin V159N mutant
To be Published
|
|
