2RLI
 
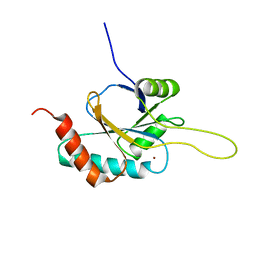 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
1EXA
 
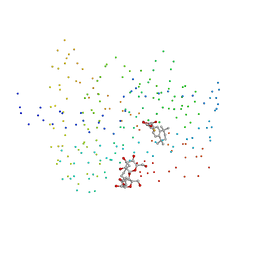 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE ACTIVE R-ENANTIOMER BMS270394. | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, R-3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1MVO
 
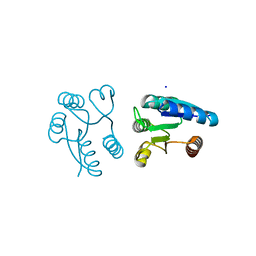 | | Crystal structure of the PhoP receiver domain from Bacillus subtilis | | Descriptor: | MANGANESE (II) ION, PhoP response regulator, SODIUM ION | | Authors: | Birck, C, Chen, Y, Hulett, F.M, Samama, J.P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-09-26 | | Release date: | 2002-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Phosphorylation Domain in PhoP Reveals a Functional Tandem Association Mediated by an Asymmetric Interface
J.BACTERIOL., 185, 2003
|
|
2G6V
 
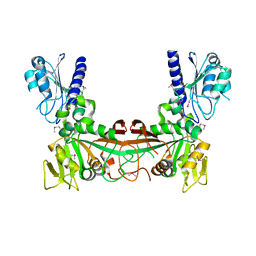 | | The crystal structure of ribD from Escherichia coli | | Descriptor: | Riboflavin biosynthesis protein ribD | | Authors: | Stenmark, P, Moche, M, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-25 | | Release date: | 2007-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Bifunctional Deaminase/Reductase RibD of the Riboflavin Biosynthetic Pathway in Escherichia coli: Implications for the Reductive Mechanism.
J.Mol.Biol., 373, 2007
|
|
2G9O
 
 | | Solution structure of the apo form of the third metal-binding domain of ATP7A protein (Menkes Disease protein) | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GA7
 
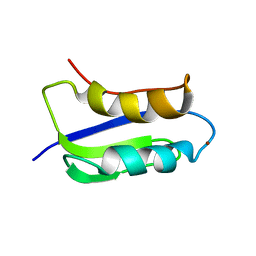 | | Solution structure of the copper(I) form of the third metal-binding domain of ATP7A protein (menkes disease protein) | | Descriptor: | COPPER (I) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, DellaMalva, N, Rosato, A, Herrmann, T, Wuthrich, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein.
J.Biol.Chem., 281, 2006
|
|
2GCF
 
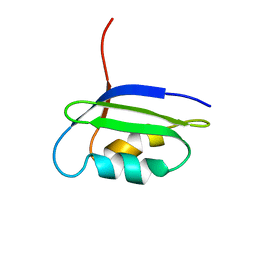 | | Solution structure of the N-terminal domain of the coppper(I) ATPase PacS in its apo form | | Descriptor: | Cation-transporting ATPase pacS | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Kandias, N.G, Spyroulias, G.A, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The delivery of copper for thylakoid import observed by NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GHF
 
 | |
2GGP
 
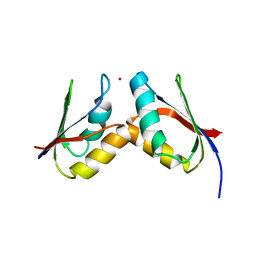 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
3LBD
 
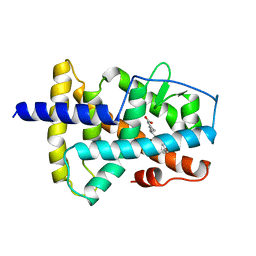 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
2KAX
 
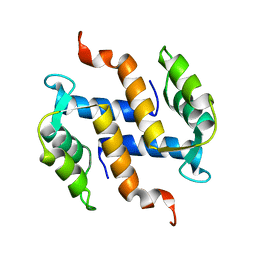 | | Solution structure and dynamics of S100A5 in the apo and Ca2+ -bound states | | Descriptor: | Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
2KAY
 
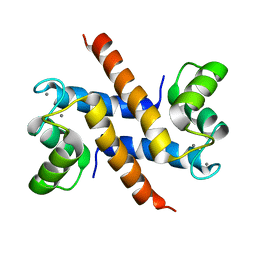 | | Solution structure and dynamics of S100A5 in the Ca2+ -bound states | | Descriptor: | CALCIUM ION, Protein S100-A5 | | Authors: | Bertini, I, Das Gupta, S, Hu, X, Karavelas, T, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-11-17 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states
J.Biol.Inorg.Chem., 14, 2009
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FCY
 
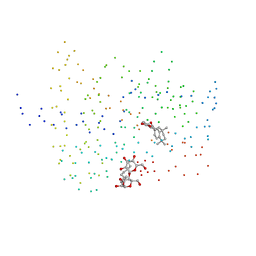 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1FCZ
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE PANAGONIST RETINOID BMS181156 | | Descriptor: | 4-[3-OXO-3-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-PROPENYL]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
4LBD
 
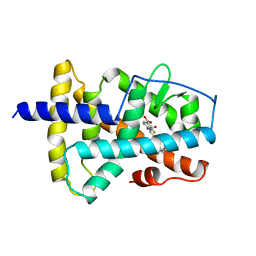 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO THE SYNTHETIC AGONIST BMS961 | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
1FCX
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARGAMMA-SELECTIVE RETINOID BMS184394 | | Descriptor: | 6-[HYDROXY-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALEN-2-YL)-METHYL]-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1FD0
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARGAMMA-SELECTIVE RETINOID SR11254 | | Descriptor: | 6-[HYDROXYIMINO-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALEN-2-YL)-METHYL]-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2002-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | C-H...O hydrogen bonds in the nuclear receptor RARgamma--a potential tool for drug selectivity.
Structure, 10, 2002
|
|
1G2N
 
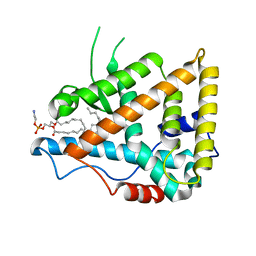 | | CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE ULTRASPIRACLE PROTEIN USP, THE ORTHOLOG OF RXRS IN INSECTS | | Descriptor: | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ULTRASPIRACLE PROTEIN | | Authors: | Billas, I.M.L, Moulinier, L, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects.
J.Biol.Chem., 276, 2001
|
|
1BON
 
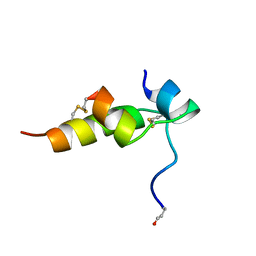 | | THREE-DIMENSIONAL STRUCTURE OF BOMBYXIN-II, AN INSULIN-RELATED BRAIN-SECRETORY PEPTIDE OF THE SILKMOTH BOMBYX MORI: COMPARISON WITH INSULIN AND RELAXIN | | Descriptor: | BOMBYXIN-II,BOMBYXIN A-2, BOMBYXIN-II,BOMBYXIN A-6 | | Authors: | Nagata, K, Hatanaka, H, Kohda, D, Inagaki, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1994-07-21 | | Release date: | 1995-01-26 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of bombyxin-II an insulin-like peptide of the silkmoth Bombyx mori: structural comparison with insulin and relaxin.
J.Mol.Biol., 253, 1995
|
|
1G50
 
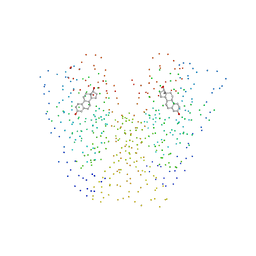 | | CRYSTAL STRUCTURE OF A WILD TYPE HER ALPHA LBD AT 2.9 ANGSTROM RESOLUTION | | Descriptor: | ESTRADIOL, ESTROGEN RECEPTOR | | Authors: | Eiler, S, Gangloff, M, Duclaud, S, Moras, D, Ruff, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-10-30 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Overexpression, Purification, and Crystal Structure of Native ER alpha LBD
Protein Expr.Purif., 22, 2001
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | Descriptor: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-11-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | Descriptor: | COPPER (II) ION, Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-21 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
1OSC
 
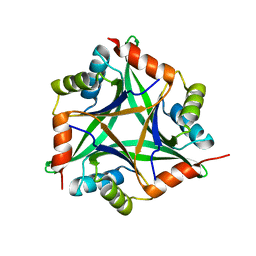 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | Descriptor: | similar to divalent cation tolerant protein CUTA | | Authors: | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
1OQ3
 
 | | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Gonnelli, L, Su, X.C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-07 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A core mutation affecting the folding properties of a soluble domain of the ATPase protein CopA from Bacillus subtilis
J.Mol.Biol., 331, 2003
|
|
