1Q33
 
 | | Crystal structure of human ADP-ribose pyrophosphatase NUDT9 | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, beta-D-glucopyranose | | Authors: | Shen, B.W, Perraud, A.L, Scharenberg, A, Stoddard, B.L. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure and Mutational Analysis of Human NUDT9
J.Mol.Biol., 332, 2003
|
|
1OX7
 
 | |
1M5X
 
 | | Crystal structure of the homing endonuclease I-MsoI bound to its DNA substrate | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-07-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
3E0L
 
 | | Computationally Designed Ammelide Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Murphy, P.M, Bolduc, J.M, Gallaher, J.L, Stoddard, B.L, Baker, D. | | Deposit date: | 2008-07-31 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Alteration of enzyme specificity by computational loop remodeling and design.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1CZ0
 
 | | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI/DNA COMPLEX LACKING CATALYTIC METAL ION | | Descriptor: | DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SODIUM ION, ... | | Authors: | Galburt, E.A, Chevalier, B, Jurica, M.S, Flick, K.E, Stoddard, B.L. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel endonuclease mechanism directly visualized for I-PpoI.
Nat.Struct.Biol., 6, 1999
|
|
1EVW
 
 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
1EVX
 
 | | APO CRYSTAL STRUCTURE OF THE HOMING ENDONUCLEASE, I-PPOI | | Descriptor: | INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SULFATE ION, ZINC ION | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
1DT0
 
 | | CLONING, SEQUENCE, AND CRYSTALLOGRAPHIC STRUCTURE OF RECOMBINANT IRON SUPEROXIDE DISMUTASE FROM PSEUDOMONAS OVALIS | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bond, C.J, Huang, J, Hajduk, R, Flick, K, Heath, P, Stoddard, B.L. | | Deposit date: | 2000-01-10 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cloning, sequence and crystallographic structure of recombinant iron superoxide dismutase from Pseudomonas ovalis.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3S1S
 
 | | Characterization and crystal structure of the type IIG restriction endonuclease BpuSI | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Shen, B.W, Xu, D, Chan, S.-H, Zheng, Y, Zhu, Y, Xu, S.-Y, Stoddard, B.L. | | Deposit date: | 2011-05-16 | | Release date: | 2011-07-13 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI.
Nucleic Acids Res., 39, 2011
|
|
1CYQ
 
 | | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI (H98A)/DNA HOMING SITE COMPLEX | | Descriptor: | 5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3', INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, MAGNESIUM ION, ... | | Authors: | Galburt, E.A, Chevalier, B, Jurica, M.S, Flick, K.E, Stoddard, B.L. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A novel endonuclease mechanism directly visualized for I-PpoI.
Nat.Struct.Biol., 6, 1999
|
|
1D7P
 
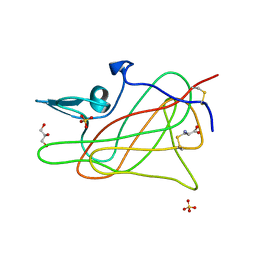 | | Crystal structure of the c2 domain of human factor viii at 1.5 a resolution at 1.5 A | | Descriptor: | COAGULATION FACTOR VIII PRECURSOR, CYSTEINE, GLYCEROL, ... | | Authors: | Pratt, K.P, Shen, B.W, Stoddard, B.L. | | Deposit date: | 1999-10-19 | | Release date: | 1999-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the C2 domain of human factor VIII at 1.5 A resolution.
Nature, 402, 1999
|
|
3R3P
 
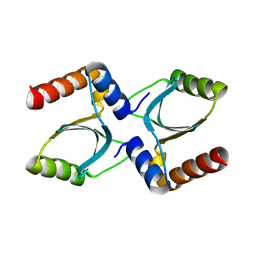 | |
3R7P
 
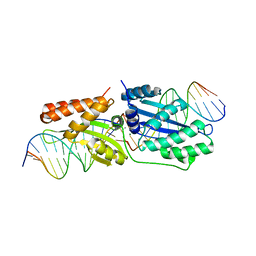 | | The crystal structure of I-LtrI | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*TP*GP*CP*TP*CP*CP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*GP*CP*AP*TP*TP*TP*G)-3'), ... | | Authors: | Mak, A.N.S, Takeuchi, R, Edgell, D.R, Stoddard, B.L. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Tapping natural reservoirs of homing endonucleases for targeted gene modification.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1G9Y
 
 | | HOMING ENDONUCLEASE I-CREI / DNA SUBSTRATE COMPLEX WITH CALCIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
1G9Z
 
 | | LAGLIDADG HOMING ENDONUCLEASE I-CREI / DNA PRODUCT COMPLEX WITH MAGNESIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*A)-3', 5'-D(P*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
3QQY
 
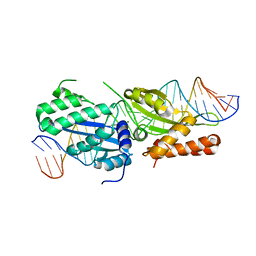 | |
3T1G
 
 | |
4OUD
 
 | | Engineered tyrosyl-tRNA synthetase with the nonstandard amino acid L-4,4-biphenylalanine | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Takeuchi, R, Mandell, D.J, Lajoie, M.J, Church, G.M, Stoddard, B.L. | | Deposit date: | 2014-02-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biocontainment of genetically modified organisms by synthetic protein design.
Nature, 518, 2015
|
|
4R6J
 
 | |
4R5D
 
 | |
4R58
 
 | |
4R5C
 
 | |
4R6G
 
 | |
4QQ8
 
 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3HYI
 
 | | Crystal structure of full-length DUF199/WhiA from Thermatoga maritima | | Descriptor: | GLYCEROL, Protein DUF199/WhiA, SODIUM ION | | Authors: | Kaiser, B.K, Clifton, M.C, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2009-06-22 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease
Structure, 17, 2009
|
|
