7JZL
 
 | |
7JZN
 
 | |
7JZM
 
 | |
7JZU
 
 | |
7KNA
 
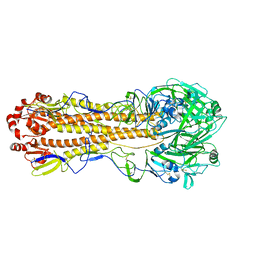 | | Localized reconstruction of the H1 A/Michigan/45/2015 ectodomain displayed at the surface of I53_dn5 nanoparticle | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,I53_dn5 | | 著者 | Acton, O.J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Quadrivalent influenza nanoparticle vaccines induce broad protection.
Nature, 592, 2021
|
|
4EFI
 
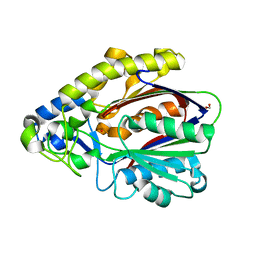 | | Crystal Structure of 3-oxoacyl-(Acyl-carrier protein) Synthase from Burkholderia Xenovorans LB400 | | 分子名称: | 3-oxoacyl-(Acyl-carrier protein) synthase, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Seattle Structural Genomics Center for Infectious Disease, Craig, T.K, Abendroth, J, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-03-29 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4F2G
 
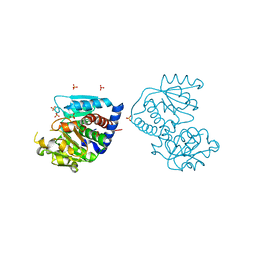 | | The Crystal Structure of Ornithine carbamoyltransferase from Burkholderia thailandensis E264 | | 分子名称: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase 1, PHOSPHATE ION | | 著者 | Craig, T.K, Fox, D, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-05-07 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
5VMR
 
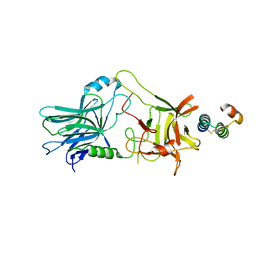 | |
5VID
 
 | |
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | 分子名称: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bernard, S.M, Wilson, I.A. | | 登録日 | 2017-04-25 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
4H4G
 
 | | Crystal Structure of (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase from Burkholderia thailandensis E264 | | 分子名称: | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | | 著者 | Craig, T.K, Edwards, T.E, Staker, B, Stewart, L, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-09-17 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
1ZO9
 
 | | Crystal Structure Of The Wild Type Heme Domain Of P450BM-3 with N-palmitoylmethionine | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | 著者 | Hegda, A, Chen, B, Tomchick, D.R, Bondlela, M, Haines, D.C, Schaffer, N, Machius, M, Graham, S.E, Peterson, J.A. | | 登録日 | 2005-05-12 | | 公開日 | 2006-08-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interactions of substrates at the surface of P450s can greatly enhance substrate potency.
Biochemistry, 46, 2007
|
|
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | 分子名称: | SULFATE ION, a novel designed pore protein, affinity purification tag | | 著者 | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | 登録日 | 2019-12-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6TJ1
 
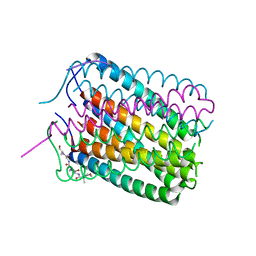 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | 分子名称: | De novo designed WSHC6, purification tag | | 著者 | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | 登録日 | 2019-11-23 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6U1S
 
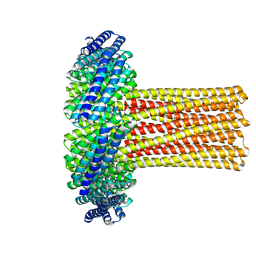 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | 分子名称: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | 著者 | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | 登録日 | 2019-08-16 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Computational Design of Transmembrane Channels
To Be Published
|
|
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | 分子名称: | TMH4C4 | | 著者 | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | 登録日 | 2020-03-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6O35
 
 | |
7UBD
 
 | |
7UBC
 
 | |
7UBF
 
 | |
7UCP
 
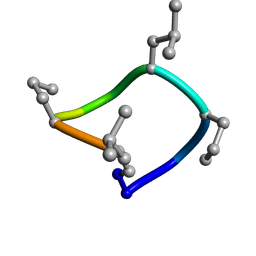 | | computationally designed macrocycle | | 分子名称: | computationally designed cyclic peptide D8.3.p2 | | 著者 | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | 登録日 | 2022-03-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-09-28 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UBE
 
 | |
7UBI
 
 | |
7UBG
 
 | |
7UBH
 
 | |
