8PS8
 
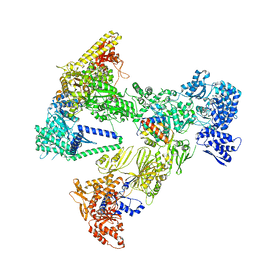 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the enoyl reductase domain (FASam sample) | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS2
 
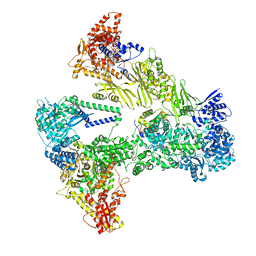 | | Asymmetric unit of the yeast fatty acid synthase with ACP at the enoyl reductase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSJ
 
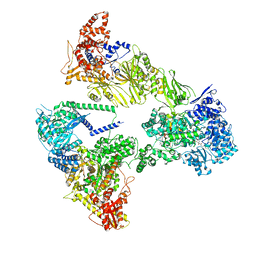 | | Asymmetric unit of the yeast fatty acid synthase in the semi rotated state with ACP at the acetyl transferase domain (FASx sample) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PS9
 
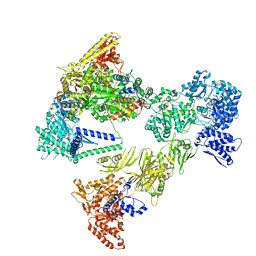 | | Asymmetric unit of the yeast fatty acid synthase in the non-rotated state with ACP at the ketosynthase domain (FASam sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSF
 
 | | Asymmetric unit of the yeast fatty acid synthase in non-rotated state with ACP at the acetyl transferase domain (FASx sample) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
8PSL
 
 | | Asymmetric unit of the yeast fatty acid synthase in the semi non-rotated state with ACP at the ketosynthase domain (FASx sample) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Singh, K, Bunzel, G, Graf, B, Yip, K.M, Stark, H, Chari, A. | | Deposit date: | 2023-07-13 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstruction of a fatty acid synthesis cycle from acyl carrier protein and cofactor structural snapshots.
Cell, 186, 2023
|
|
5LZA
 
 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZF
 
 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZD
 
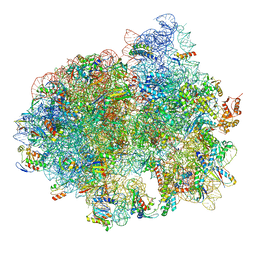 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZC
 
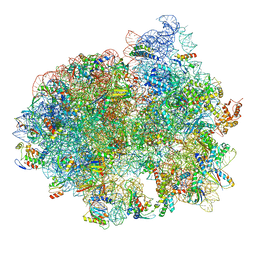 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the codon reading state (CR) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZE
 
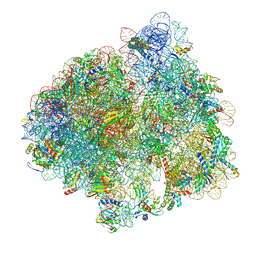 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZB
 
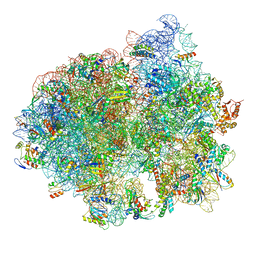 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
6H60
 
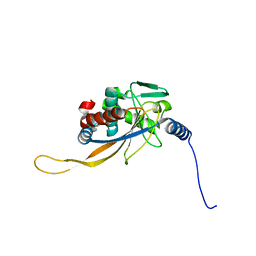 | | pseudo-atomic structural model of the E3BP component of the human pyruvate dehydrogenase multienzyme complex | | Descriptor: | Pyruvate dehydrogenase protein X component, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6Z9F
 
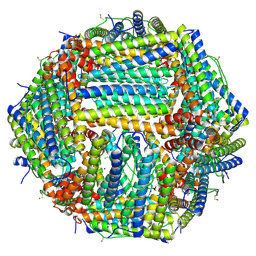 | | 1.56 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.56 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6Z6U
 
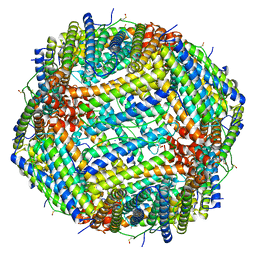 | | 1.25 A structure of human apoferritin obtained from Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-05-29 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.25 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6Z9E
 
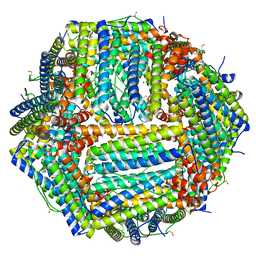 | | 1.55 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
7A6B
 
 | | 1.33 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.33 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
7A6A
 
 | | 1.15 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.15 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-17 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MT5
 
 | | Isolated Ring domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissman, F, Royappa, G, Schulman, B, Jarvis, M, Vanderlinden, R, Frye, J.J, Qiao, R, Petzold, G, Peters, J, Stark, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
5KHU
 
 | | Model of human Anaphase-promoting complex/Cyclosome (APC15 deletion mutant), in complex with the Mitotic checkpoint complex (APC/C-CDC20-MCC) based on cryo EM data at 4.8 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Yamaguchi, M, VanderLinden, R, Dube, P, Stark, H, Schulman, B. | | Deposit date: | 2016-06-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
5KHR
 
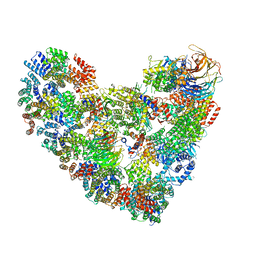 | | Model of human Anaphase-promoting complex/Cyclosome complex (APC15 deletion mutant) in complex with the E2 UBE2C/UBCH10 poised for ubiquitin ligation to substrate (APC/C-CDC20-substrate-UBE2C) | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | VanderLinden, R, Yamaguchi, M, Dube, P, Haselbach, D, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
5L9U
 
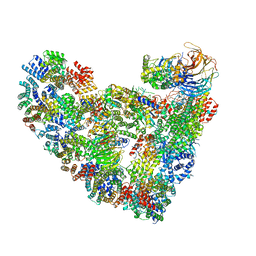 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
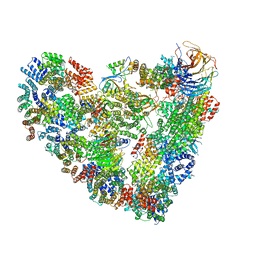 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2016-11-02 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
