2OO6
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 | | Descriptor: | Putative L-alanine-DL-glutamate epimerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Sridhar, V, Freeman, J, Smyth, L, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400
To be Published
|
|
2OZ8
 
 | | Crystal structure of putative mandelate racemase from Mesorhizobium loti | | Descriptor: | Mll7089 protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of putative mandelate racemase from Mesorhizobium loti
To be Published
|
|
2OO2
 
 | | Crystal structure of protein AF1782 from Archaeoglobus fulgidus, Pfam DUF357 | | Descriptor: | Hypothetical protein AF_1782 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Adams, J, Sridhar, V, Smyth, L, Freeman, J, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical AF_1782 protein from Archaeoglobus fulgidus
To be Published
|
|
2P1J
 
 | | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima | | Descriptor: | DNA polymerase III polC-type | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Izuka, M, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima
To be Published
|
|
2P1G
 
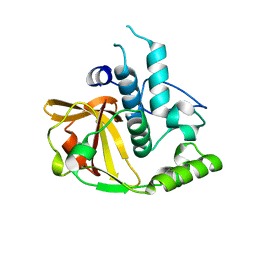 | | Crystal structure of a putative xylanase from Bacteroides fragilis | | Descriptor: | Putative xylanase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative xylanase from Bacteroides fragilis
To be Published
|
|
2Q01
 
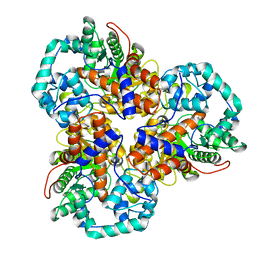 | | Crystal structure of glucuronate isomerase from Caulobacter crescentus | | Descriptor: | POTASSIUM ION, Uronate isomerase | | Authors: | Patskovsky, Y, Bonanno, J, Sridhar, V, Sauder, J.M, Freeman, J, Powell, A, Koss, J, Groshong, C, Gheyi, T, Wasserman, S.R, Raushel, F, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Glucuronate Isomerase from Caulobacter crescentus.
To be Published
|
|
1AXQ
 
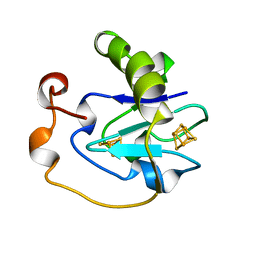 | | FERRICYANIDE OXIDIZED FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of ferricyanide-oxidized [Fe-S] clusters in Azotobacter vinelandii ferredoxin I.
J.Biol.Inorg.Chem., 3, 1998
|
|
1FOC
 
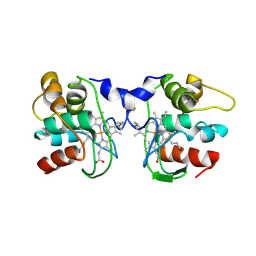 | | Cytochrome C557: improperly folded thermus thermophilus C552 | | Descriptor: | CYTOCHROME RC557, HEME C | | Authors: | McRee, D.E, Williams, P.A, Fee, J.A, Bren, K.L. | | Deposit date: | 2000-08-27 | | Release date: | 2000-11-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recombinant cytochrome rC557 obtained from Escherichia coli cells expressing a truncated Thermus thermophilus cycA gene. Heme inversion in an improperly matured protein
J.Biol.Chem., 276, 2001
|
|
2FNO
 
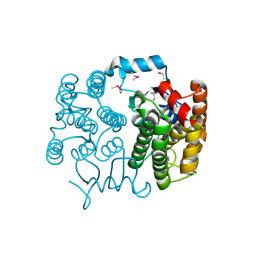 | |
4MSA
 
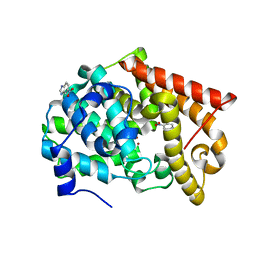 | | Crystal structure of PDE10A2 with fragment ZT0449 (5-nitro-1H-benzimidazole) | | Descriptor: | 5-nitro-1H-benzimidazole, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
1G3O
 
 | | CRYSTAL STRUCTURE OF V19E MUTANT OF FERREDOXIN I | | Descriptor: | 7FE FERREDOXIN I, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D, Burgess, B.K, Bonagura, C.A, Jung, Y.S. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Azotobacter vinelandii ferredoxin I: a sequence and structure comparison approach to alteration of [4Fe-4S]2+/+ reduction potential
J.Biol.Chem., 277, 2002
|
|
1G6B
 
 | | CRYSTAL STRUCTURE OF P47S MUTANT OF FERREDOXIN I | | Descriptor: | 7FE FERREDOXIN I, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D, Burgess, B.K, Bonagura, C.A, Jung, Y.S. | | Deposit date: | 2000-11-03 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azotobacter vinelandii ferredoxin I: a sequence and structure comparison approach to alteration of [4Fe-4S]2+/+ reduction potential.
J.Biol.Chem., 277, 2002
|
|
2OG9
 
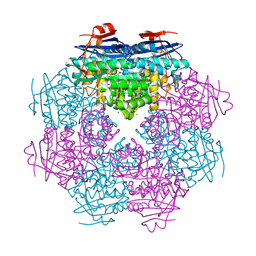 | |
1N6B
 
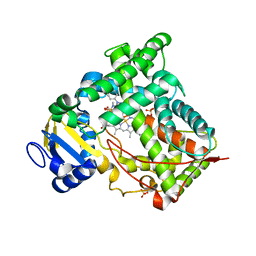 | | Microsomal Cytochrome P450 2C5/3LVdH Complex with a dimethyl derivative of sulfaphenazole | | Descriptor: | 4-METHYL-N-METHYL-N-(2-PHENYL-2H-PYRAZOL-3-YL)BENZENESULFONAMIDE, Cytochrome P450 2C5, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wester, M.R, Johnson, E.F, Marques-Soares, C, Dansette, P.M, Mansuy, D, Stout, C.D. | | Deposit date: | 2002-11-09 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Substrate Complex of Mammalian Cytochrome P450 2C5 at 2.3 A Resolution: Evidence
for Multiple Substrate Binding Modes
Biochemistry, 42, 2003
|
|
1PFF
 
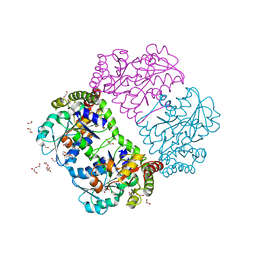 | | Crystal Structure of Homocysteine alpha-, gamma-lyase at 1.8 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, methionine gamma-lyase | | Authors: | Allen, T.W, Sridhar, V, Prasad, S.G, Han, Q, Xu, M, Tan, Y, Hoffman, R.M, Ramaswamy, S. | | Deposit date: | 2003-05-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: |
|
|
1PG8
 
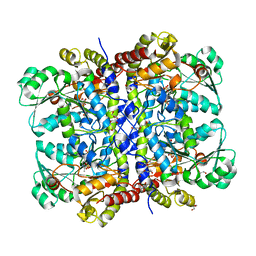 | | Crystal Structure of L-methionine alpha-, gamma-lyase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Allen, T.W, Sridhar, V, Prasad, G.S, Han, Q, Xu, M, Tan, Y, Hoffman, R.M, Ramaswamy, S. | | Deposit date: | 2003-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: |
To Be Published
|
|
2P5Z
 
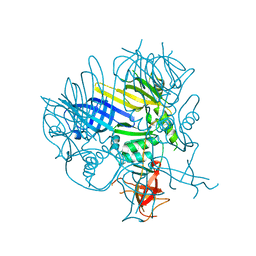 | | The E. coli c3393 protein is a component of the type VI secretion system and exhibits structural similarity to T4 bacteriophage tail proteins gp27 and gp5 | | Descriptor: | Type VI secretion system component | | Authors: | Ramagopal, U.A, Bonanno, J.B, Sridhar, V, Lau, C, Toro, R, Gheyi, T, Maletic, M, Freeman, J.C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
2PMQ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
