4GLK
 
 | | Structure and activity of AbiQ, a lactococcal anti-phage endoribonuclease belonging to the type-III toxin-antitoxin system | | Descriptor: | AbiQ, GLYCEROL | | Authors: | Samson, J, Spinelli, S, Cambillau, C, Moineau, S. | | Deposit date: | 2012-08-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type III toxin-antitoxin system.
Mol.Microbiol., 87, 2013
|
|
5M2J
 
 | | Complex between human TNF alpha and Llama VHH2 | | Descriptor: | Anti-(ED-B) scFV, Tumor necrosis factor | | Authors: | Cambillau, C, Spinelli, S, Desmyter, A, de Haard, H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions.
Front Immunol, 8, 2017
|
|
5M2M
 
 | | Complex between human TNF alpha and Llama VHH3 | | Descriptor: | Llama nanobody VHH3, Tumor necrosis factor | | Authors: | Cambillau, C, Spinelli, S, Desmyter, A, de Haard, H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions.
Front Immunol, 8, 2017
|
|
5LY8
 
 | | Structure of the CBM2 module of Lactobacillus casei BL23 phage J-1 evolved Dit. | | Descriptor: | MAGNESIUM ION, Tail component | | Authors: | Cambillau, C, Spinelli, S, Dieterle, M.-E, Piuri, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: a novel type of anti-receptor widespread among lactic acid bacteria phages.
Mol. Microbiol., 104, 2017
|
|
5M2Y
 
 | | Structure of TssK C-terminal domain from E. coli T6SS | | Descriptor: | TssK C | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A, Legrand, P. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
5M2I
 
 | | Structure of human Tumor Necrosis Factor (TNF) in complex with the Llama VHH1 | | Descriptor: | Tumor necrosis factor, VHH1 | | Authors: | Cambillau, C, Spinelli, S, Desmyter, A, de Haard, H. | | Deposit date: | 2016-10-13 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Bivalent Llama Single-Domain Antibody Fragments against Tumor Necrosis Factor Have Picomolar Potencies due to Intramolecular Interactions.
Front Immunol, 8, 2017
|
|
5M2W
 
 | | Structure of nanobody nb18 raised against TssK from E. coli T6SS | | Descriptor: | Llama nanobody nb8 against TssK from T6SS, SULFATE ION | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
4HEM
 
 | | Llama vHH-02 binder of ORF49 (RBP) from lactococcal phage TP901-1 | | Descriptor: | Anti-baseplate TP901-1 Llama vHH 02, BPP | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HEP
 
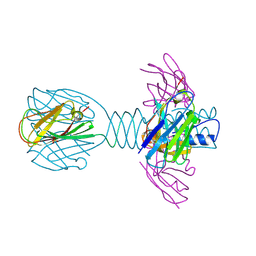 | | Complex of lactococcal phage TP901-1 with a llama vHH (vHH17) binder (nanobody) | | Descriptor: | BPP, SULFATE ION, vHH17 domain | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5MWN
 
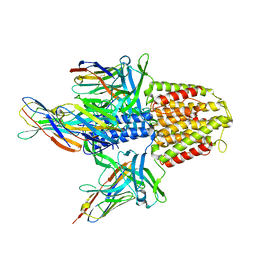 | | Structure of the EAEC T6SS component TssK N-terminal domain in complex with llama nanobodies nbK18 and nbK27 | | Descriptor: | PHOSPHATE ION, Type VI secretion protein, llama nanobody raised against TssK, ... | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Legrand, P. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
4HKH
 
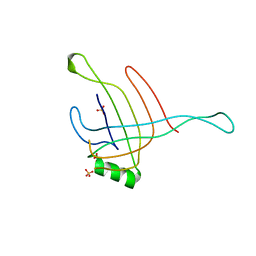 | | Structure of the Hcp1 protein from E. coli EAEC 042 pathovar, mutants N93W-S158W | | Descriptor: | Putative type VI secretion protein, SULFATE ION | | Authors: | Douzi, B, Spinelli, S, Derrez, E, Blangy, S, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Hcp1 protein from E. coli EAEC 042 pathovar, mutants N93W-S158W
To be Published
|
|
1W8G
 
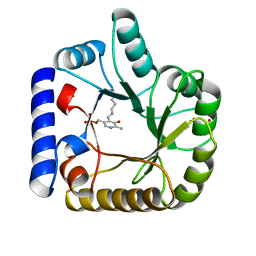 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
4IOS
 
 | | Structure of phage TP901-1 RBP (ORF49) in complex with nanobody 11. | | Descriptor: | BPP, GLYCEROL, Llama nanobody 11 | | Authors: | Desmyter, A, Farenc, C, Mahony, J, Spinelli, S, Bebeacua, C, Blangy, S, Veesler, D, van Sinderen, D, Cambillau, C. | | Deposit date: | 2013-01-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3U6X
 
 | | Phage TP901-1 baseplate tripod | | Descriptor: | BPP, BROMIDE ION, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NNZ
 
 | |
3UH8
 
 | | N-terminal domain of phage TP901-1 ORF48 | | Descriptor: | ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4O9H
 
 | | Structure of Interleukin-6 in complex with a Camelid Fab fragment | | Descriptor: | Heavy Chain of the Camelid Fab fragment 61H7, Interleukin-6, Light Chain of the Camelid Fab fragment 61H7 | | Authors: | Klarenbeek, A, Blanchetot, C, Schragel, G, Sadi, A.S, Ongenae, N, Hemrika, W, Wijdenes, J, Spinelli, S, Desmyter, A, Cambillau, C, Hultberg, A, Kretz-rommel, A, Dreier, T, De haard, H.J.W, Roovers, R.C. | | Deposit date: | 2014-01-02 | | Release date: | 2015-04-15 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Combining residues of naturally-occurring Camelid somatic affinity variants yields ultra-potent human therapeutic IL-6 antibodies
To be Published
|
|
2A9M
 
 | |
2A9N
 
 | | A Mutation Designed to Alter Crystal Packing Permits Structural Analysis of a Tight-binding Fluorescein-scFv complex | | Descriptor: | 4-(2,7-DIFLUORO-6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)ISOPHTHALIC ACID, fluorescein-scfv | | Authors: | Cambillau, C, Spinelli, S, Honegger, A, Pluckthun, A. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation designed to alter crystal packing permits structural analysis of a tight-binding fluorescein-scFv complex.
Protein Sci., 14, 2005
|
|
1P28
 
 | | The crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae in complex with a component of the pheromonal blend: 3-hydroxy-butan-2-one. | | Descriptor: | R,3-HYDROXYBUTAN-2-ONE, S,3-HYDROXYBUTAN-2-ONE, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
1ORG
 
 | | The crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae reveals a new mechanism of pheromone binding | | Descriptor: | GLYCEROL, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-03-13 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
1OW4
 
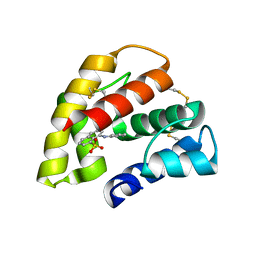 | | Crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae in complex with the fluorescent reporter ANS (1-anilinonaphtalene-8-sulfonic acid), | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-03-28 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
1QFW
 
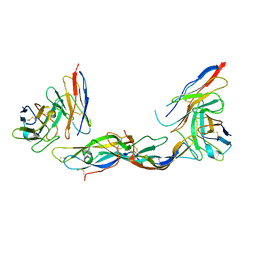 | | TERNARY COMPLEX OF HUMAN CHORIONIC GONADOTROPIN WITH FV ANTI ALPHA SUBUNIT AND FV ANTI BETA SUBUNIT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (ANTI ALPHA SUBUNIT) (HEAVY CHAIN), ANTIBODY (ANTI ALPHA SUBUNIT) (LIGHT CHAIN), ... | | Authors: | Tegoni, M, Spinelli, S, Cambillau, C. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a ternary complex between human chorionic gonadotropin (hCG) and two Fv fragments specific for the alpha and beta-subunits.
J.Mol.Biol., 289, 1999
|
|
1NZJ
 
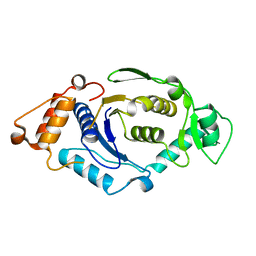 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
1KXT
 
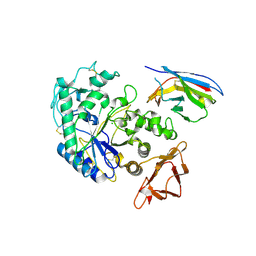 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
