6DEU
 
 | | Human caspase-6 A109T | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
6DEV
 
 | | Human caspase-6 E35K | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
6DJX
 
 | | Crystal Structure of pParkin-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DJW
 
 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7MP8
 
 | |
7MP9
 
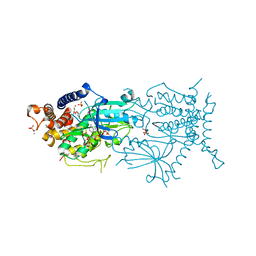 | | Crystal structure of the cytosolic domain of Tribolium castaneum PINK1 phosphorylated at Ser205 in complex with ADP analog | | Descriptor: | AMP PHOSPHORAMIDATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Rasool, S, Veyron, S, Trempe, J.F. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of PINK1 activation by autophosphorylation and insights into assembly on the TOM complex.
Mol.Cell, 82, 2022
|
|
8XUJ
 
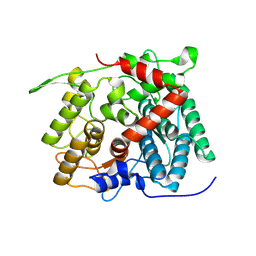 | |
8XUL
 
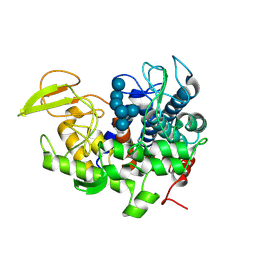 | | Structure of beta-1,2-glucanase from Xanthomonas campestris pv. campestris (beta-1,2-glucoheptasaccharide complex)-E239Q mutant | | Descriptor: | XCC2207, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Nakajima, M, Motouchi, S, Nakai, H. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | beta-1,2-Glucanase superfamily identified by sequential, functional and structural analyses
To Be Published
|
|
8XUK
 
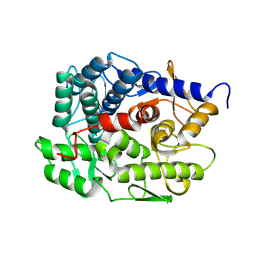 | | Structure of beta-1,2-glucanase from Photobacterium gaetbulicola (PgSGL3, ligand-free) | | Descriptor: | CHLORIDE ION, H744_1c0222 | | Authors: | Nakajima, M, Motouchi, S, Nakai, H. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | beta-1,2-Glucanase superfamily identified by sequential, functional and structural analyses
To Be Published
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | Descriptor: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.61 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
