3WVZ
 
 | | Crystal structure of Hikeshi, a new nuclear transport receptor of Hsp70 | | Descriptor: | Protein Hikeshi | | Authors: | Song, J, Kose, S, Watanabe, A, Son, S.Y, Choi, S, Hong, R.H, Yamashita, E, Park, I.Y, Imamoto, N, Lee, S.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70s
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WW0
 
 | | Crystal structure of F97A mutant, a new nuclear transport receptor of Hsp70 | | Descriptor: | Protein Hikeshi | | Authors: | Song, J, Kose, S, Watanabe, A, Son, S.Y, Choi, S, Hong, R.H, Yamashita, E, Park, I.Y, Imamoto, N, Lee, S.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of Hikeshi, a new nuclear transport receptor of Hsp70s
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7V8V
 
 | | Crystal structure of PsEst3 S128A mutant | | Descriptor: | esterase | | Authors: | Son, J, Kim, H, Kim, H.W. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
7V8U
 
 | | Crystal structure of PsEst3 wild-type | | Descriptor: | Esterase, NITROBENZENE, SULFATE ION | | Authors: | Son, J, Kim, H, Kim, H.W. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
7V8X
 
 | |
7V8W
 
 | | Crystal structure of PsEst3 S128A variant complexed with malonate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MALONIC ACID, ... | | Authors: | Son, J, Kim, H, Kim, H.W. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
5CZY
 
 | | Crystal structure of LegAS4 | | Descriptor: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | Authors: | Son, J, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
2G2B
 
 | | NMR structure of the human allograft inflammatory factor 1 | | Descriptor: | Allograft inflammatory factor 1 | | Authors: | Song, J, Tyler, R.C, Newman, C.L, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-15 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human allograft inflammatory factor 1
To be published
|
|
2GW6
 
 | |
5ZGA
 
 | | Crystal Structure of Triosephosphate isomerase SAD deletion and N115A mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZG5
 
 | | Crystal Structure of Triosephosphate isomerase SADsubAAA mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZFX
 
 | | Crystal Structure of Triosephosphate isomerase from Opisthorchis viverrini | | Descriptor: | MAGNESIUM ION, Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
5ZG4
 
 | | Crystal Structure of Triosephosphate isomerase SAD deletion mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
8WZU
 
 | |
8FYH
 
 | | G4 RNA-mediated PRC2 dimer | | Descriptor: | G4 RNA, Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Song, J, Kasinath, V. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inactivation of PRC2 by G-quadruplex RNA.
Science, 381, 2023
|
|
6YMY
 
 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
6YMX
 
 | | CIII2/CIV respiratory supercomplex from Saccharomyces cerevisiae | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
4ZDS
 
 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | Descriptor: | Protein ETHYLENE INSENSITIVE 3 | | Authors: | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
2W0D
 
 | | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? A Case Study of Metalloproteinases. | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Isaksson, J, Nystrom, S, Derbyshire, D.J, Wallberg, H, Agback, T, Kovacs, H, Bertini, I, Felli, I.C. | | Deposit date: | 2008-08-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? a Case Study of Metalloproteinases.
J.Med.Chem., 52, 2009
|
|
1FFT
 
 | | The structure of ubiquinol oxidase from Escherichia coli | | Descriptor: | COPPER (II) ION, HEME O, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abramson, J, Riistama, S, Larsson, G, Jasaitis, A, Svensson-Ek, M, Puustinen, A, Iwata, S, Wikstrom, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site.
Nat.Struct.Biol., 7, 2000
|
|
3PTA
 
 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PT9
 
 | | Crystal structure of mouse DNMT1(731-1602) in the free state | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PT6
 
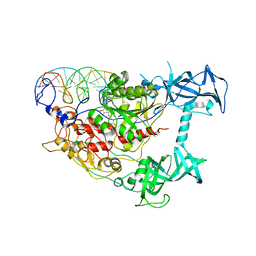 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
4CA3
 
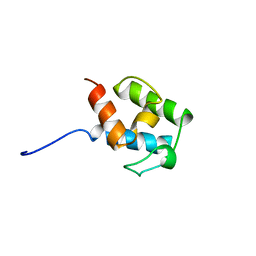 | | SOLUTION STRUCTURE OF STREPTOMYCES VIRGINIAE VIRA ACP5B | | Descriptor: | HYBRID POLYKETIDE SYNTHASE-NON RIBOSOMAL PEPTIDE SYNTHETASE | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights Into the Function of Trans-Acyl Transferase Polyketide Synthases from the Saxs Structure of a Complete Module.
Chem.Sci., 2014
|
|
1SPF
 
 | |
