3O97
 
 | | Crystal Structure of the complex of C-lobe of lactoferrin with indole acetic acid at 2.68 A Resolution | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Sinha, M, Bhushan, A, Vikram, G, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of the complex of C-lobe of lactoferrin with indole acetic acid at 2.68 A Resolution
To be Published
|
|
2AYW
 
 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4ZC1
 
 | |
4ZGB
 
 | | Structure of untreated lipase from Thermomyces lanuginosa at 2.3 A resolution | | Descriptor: | Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-04-22 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
4ZT8
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a pyrimidine base, cytosine at 1.98 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOPYRIMIDIN-2(1H)-ONE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-14 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4ZU0
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine monophosphate at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4ZZ6
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine triphosphate at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5B6P
 
 | | Structure of the dodecameric type-II dehydrogenate dehydratase from Acinetobacter baumannii at 2.00 A resolution | | Descriptor: | 3-dehydroquinate dehydratase, SULFATE ION | | Authors: | Kumar, M, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding studies and structure determination of the recombinantly produced type-II 3-dehydroquinate dehydratase from Acinetobacter baumannii.
Int. J. Biol. Macromol., 94, 2017
|
|
5CFK
 
 | | Crystal structure of Proliferating Cell Nuclear Antigen from Leishmania donovani at 3.2 A resolution | | Descriptor: | Proliferating cell nuclear antigen,Proliferating cell nuclear antigen | | Authors: | Shukla, P.K, Yadav, S.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Proliferating Cell Nuclear Antigen from Leishmania donovani at 3.2 A resolution
To Be Published
|
|
5CSO
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleoside, cytidine at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5CST
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine diphosphate at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5E0B
 
 | | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, N-acetyl-beta-muramic acid, ... | | Authors: | Sharma, P, Yamini, S, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution
To Be Published
|
|
5E0A
 
 | | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A
To Be Published
|
|
5YL5
 
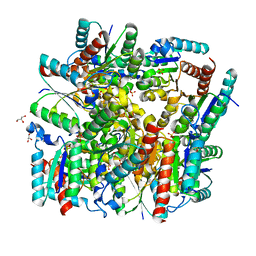 | | Crystal structure of dodecameric Dehydroquinate dehydratase from Acinetobacter baumannii at 1.9A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, GLYCEROL, ... | | Authors: | Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dodecameric Dehydroquinate dehydratase from Acinetobacter baumannii at 1.9A resolution
To Be Published
|
|
5YL8
 
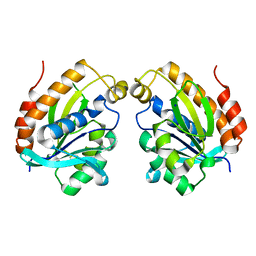 | | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution
To Be Published
|
|
1P7V
 
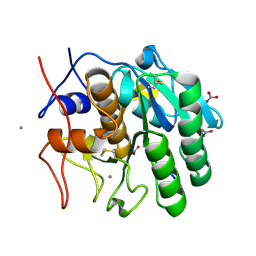 | | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Kaur, P, Chandra, V, Banumathi, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution
To be published
|
|
1OZ6
 
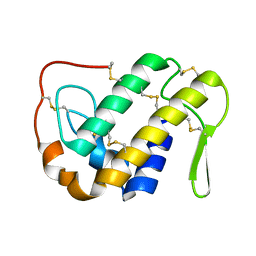 | | X-ray structure of acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) with a potent platelet aggregation inhibitory activity | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Jasti, J, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-08 | | Release date: | 2003-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from Indian saw-scaled viper (Echis carinatus) at 2.6 A resolution reveals a novel intermolecular interaction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OZ7
 
 | | Crystal structure of Echicetin from the venom of Indian saw-scaled viper (Echis carinatus) at 2.4 resolution | | Descriptor: | echicetin A-chain, echicetin B-chain | | Authors: | Jasti, J, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-08 | | Release date: | 2003-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of echicetin from Echis carinatus (Indian saw-scaled viper) at 2.4A resolution.
J.Mol.Biol., 335, 2004
|
|
4MPK
 
 | | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, GLYCEROL | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution
To be Published
|
|
1PFG
 
 | | Strategy to design inhibitors: Structure of a complex of Proteinase K with a designed octapeptide inhibitor N-Ac-Pro-Ala-Pro-Phe-DAla-Ala-Ala-Ala-NH2 at 2.5A resolution | | Descriptor: | N-Ac-PAPFAAAA-NH2, Proteinase K | | Authors: | Saxena, A.K, Singh, T.P, Peters, K, Fittkau, S, Betzel, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Strategy to design peptide inhibitors: structure of a complex of proteinase K with a designed octapeptide inhibitor N-Ac-Pro-Ala-Pro-Phe-DAla-Ala-Ala-Ala-NH2 at 2.5 A resolution.
Protein Sci., 5, 1996
|
|
1PEK
 
 | | STRUCTURE OF THE COMPLEX OF PROTEINASE K WITH A SUBSTRATE-ANALOGUE HEXA-PEPTIDE INHIBITOR AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | D-DAL-ALA-NH2, PEPTIDE PRO-ALA-PRO-PHE, PROTEINASE K | | Authors: | Betzel, C, Singh, T.P, Visanji, M, Peters, K, Fittkau, S, Saenger, W, Wilson, K.S. | | Deposit date: | 1993-01-19 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution.
J.Biol.Chem., 268, 1993
|
|
1PJ8
 
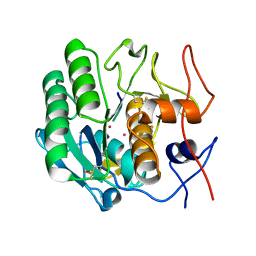 | | Structure of a ternary complex of proteinase K, mercury and a substrate-analogue hexapeptide at 2.2 A resolution | | Descriptor: | 6-residue peptide (N-Ac-PAPFPA-NH2), MERCURY (II) ION, Proteinase K | | Authors: | Saxena, A.K, Singh, T.P, Peters, K, Fittkau, S, Visanji, M, Wilson, K.S, Betzel, C. | | Deposit date: | 2003-06-02 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ternary complex of proteinase K, mercury, and a substrate-analogue hexa-peptide at 2.2 A resolution
Proteins, 25, 1996
|
|
4N6P
 
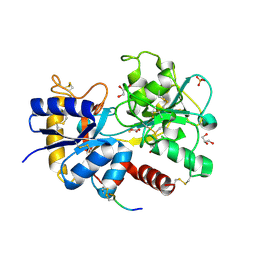 | | Crystal Structure of C-lobe of Bovine lactoferrin complexed with meclofenamic acid at 1.4 A resolution | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gautam, L, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-14 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structre of C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH MECLOFENAMIC ACID AT 1.4 A RESOLUTION
TO BE PUBLISHED
|
|
4O0O
 
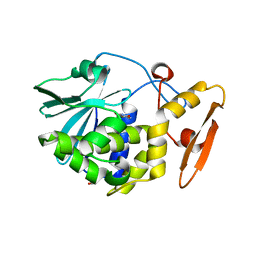 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with 5-fluorouracil at 2.59 A resolution
To be Published
|
|
4NJB
 
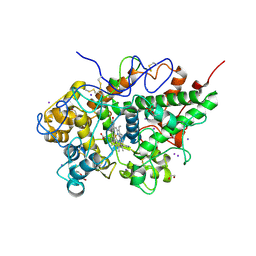 | | Crystal structure of the complex of lactoperoxidase from bovine with 3,3-oxydipyridine at 2.31 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Sirohi, H.V, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-11-09 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the complex of lactoperoxidase from bovine with 3,3-oxydipyridine at 2.31 A resolution
To be Published
|
|
