1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | Authors: | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | Deposit date: | 2005-06-01 | | Release date: | 2006-06-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
5VSG
 
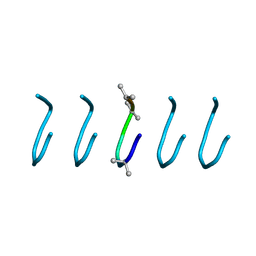 | | Fibrils of the super helical repeat peptide, SHR-FF, grown at elevated temperature | | Descriptor: | Super Helical Repeat Peptide SHR-FF | | Authors: | Mondal, S, Sawaya, M.R, Eisenberg, D.S, Gazit, E. | | Deposit date: | 2017-05-11 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Transition of Metastable Cross-alpha Crystals into Cross-beta Fibrils by beta-Turn Flipping.
J.Am.Chem.Soc., 141, 2019
|
|
3ZQW
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-01-11 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1Y9A
 
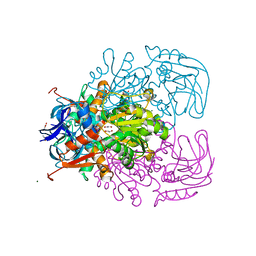 | | Alcohol Dehydrogenase from Entamoeba histolotica in complex with cacodylate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CACODYLATE ION, ... | | Authors: | Shimon, L.J, Peretz, M, Goihberg, E, Burstein, Y, Frolow, F. | | Deposit date: | 2004-12-15 | | Release date: | 2006-01-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of alcohol dehydrogenase from Entamoeba histolytica.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1QZN
 
 | | Crystal Structure Analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus | | Descriptor: | cellulosomal scaffoldin adaptor protein B | | Authors: | Frolow, F, Noach, I, Rosenheck, S, Lamed, R, Qi, X, Shimon, L.J.W, Bayer, E.A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements.
J.Mol.Biol., 348, 2005
|
|
2ZF9
 
 | |
3FNK
 
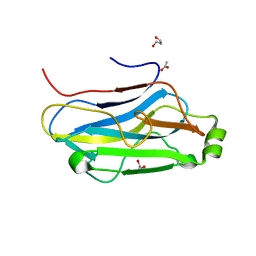 | | Crystal structure of the second type II cohesin module from the cellulosomal adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2008-12-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Intermodular Linker Flexibility Revealed from Crystal Structures of Adjacent Cellulosomal Cohesins of Acetivibrio cellulolyticus
J.Mol.Biol., 391, 2009
|
|
3ZU8
 
 | | STRUCTURE OF CBM3B OF MAJOR SCAFFOLDIN SUBUNIT SCAA FROM ACETIVIBRIO CELLULOLYTICUS DETERMINED ON THE NIKEL ABSORPTION EDGE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ZUC
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus determined from the crystals grown in the presence of Nickel | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-07-18 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3D07
 
 | |
3D09
 
 | |
3D0A
 
 | |
4B96
 
 | | Family 3b carbohydrate-binding module from the biomass sensoring system of Clostridium clariflavum | | Descriptor: | CALCIUM ION, CELLULOSE BINDING DOMAIN-CONTAINING PROTEIN, CHLORIDE ION | | Authors: | Yaniv, O, Reddy, Y.H.K, Yoffe, H, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-02 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structure of Cbm3B from the Biomass Sensoring System of Clostridium Clarifalvum
To be Published
|
|
