6X6O
 
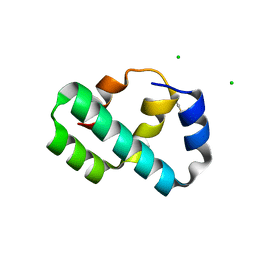 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | 分子名称: | CHLORIDE ION, Protein spackle | | 著者 | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6XC1
 
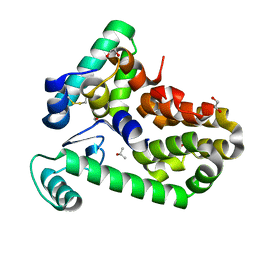 | | Crystal structure of bacteriophage T4 spackle and lysozyme in orthorhombic form | | 分子名称: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Lysozyme, ... | | 著者 | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | 登録日 | 2020-06-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6XC0
 
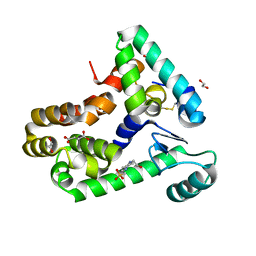 | | Crystal structure of bacteriophage T4 spackle and lysozyme in monoclinic form | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | 登録日 | 2020-06-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6DRT
 
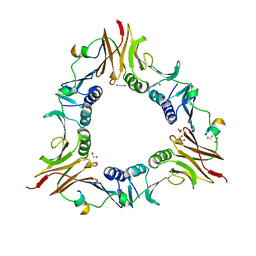 | |
6DT1
 
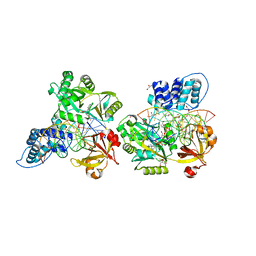 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2018-06-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
6NFK
 
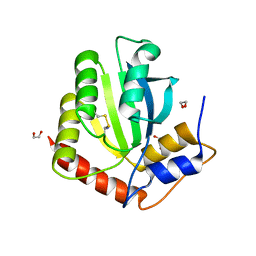 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | 分子名称: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | 著者 | Shi, K, Orellana, K, Aihara, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | 分子名称: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | 著者 | Shi, K, Orellana, K, Aihara, H. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.731 Å) | | 主引用文献 | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
1DVL
 
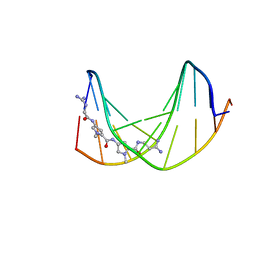 | |
6CWJ
 
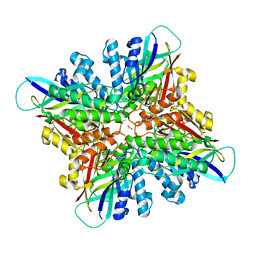 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with 1,3-Acetone Dicarboxylic Acid | | 分子名称: | 1,3-PROPANDIOL, 3-oxopentanedioic acid, ACETATE ION, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2018-03-30 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.253 Å) | | 主引用文献 | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6DHJ
 
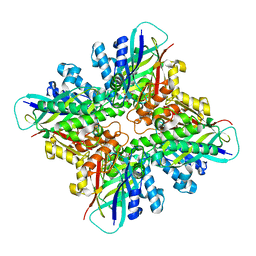 | |
6NFM
 
 | |
1R3Z
 
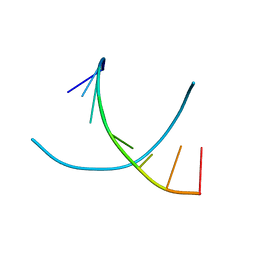 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | 分子名称: | 5'-D(*GP*(5CM)P*GP*(5CM)P*GP*CP*GP*C)-3' | | 著者 | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | 登録日 | 2003-10-03 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R41
 
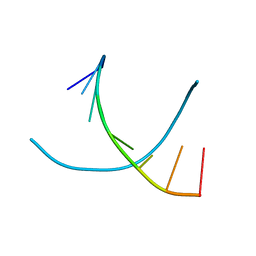 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | 分子名称: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | 著者 | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | 登録日 | 2003-10-03 | | 公開日 | 2003-12-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5CQK
 
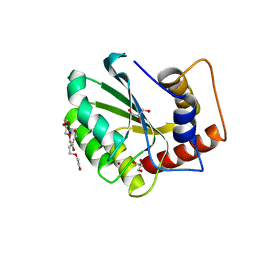 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | 分子名称: | DNA dC->dU-editing enzyme APOBEC-3B, GLYCEROL, SODIUM ION, ... | | 著者 | Shi, K, Kurahashi, K, Aihara, H. | | 登録日 | 2015-07-21 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQI
 
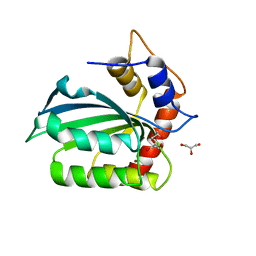 | |
5CQH
 
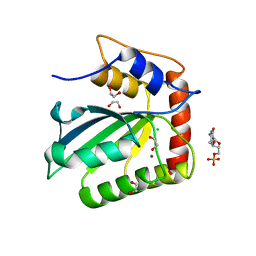 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, ... | | 著者 | Shi, K, Kurahashi, K, Aihara, H. | | 登録日 | 2015-07-21 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQD
 
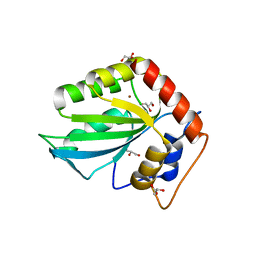 | |
438D
 
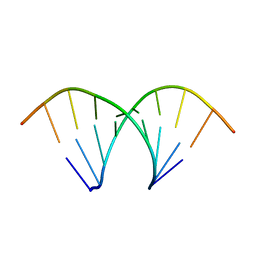 | |
4E10
 
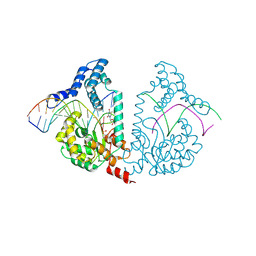 | | Protelomerase tela Y201A covalently complexed with substrate DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*AP*(BRU)P*AP*AP*CP*AP*AP*(BRU)P*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*(BRU)P*(BRU)P*GP*(BRU)P*(BRU)P*AP*(BRU)P*(BRU)P*AP*(BRU)P*G)-3'), Protelomerase, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2012-03-05 | | 公開日 | 2013-02-13 | | 実験手法 | X-RAY DIFFRACTION (2.506 Å) | | 主引用文献 | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4DT8
 
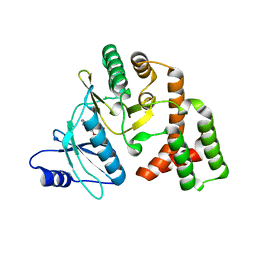 | |
4DTA
 
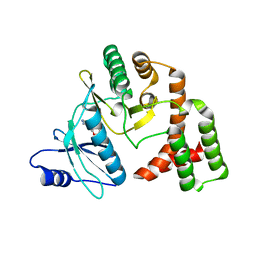 | |
4DT9
 
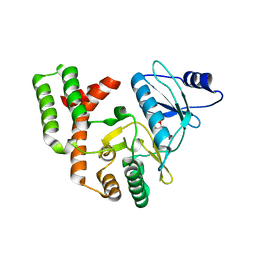 | |
4DWP
 
 | | SeMet protelomerase tela covalently complexed with substrate DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*T)-3'), Protelomerase, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2012-02-26 | | 公開日 | 2013-02-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4DTB
 
 | |
4E0G
 
 | | Protelomerase tela/DNA hairpin product/vanadate complex | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*AP*TP*AP*AP*CP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*AP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*AP*TP*G)-3'), Protelomerase, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2012-03-03 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
