6B3A
 
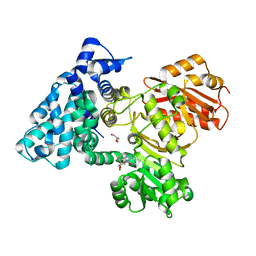 | |
6B3B
 
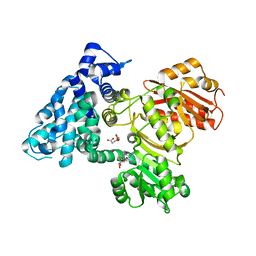 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6B39
 
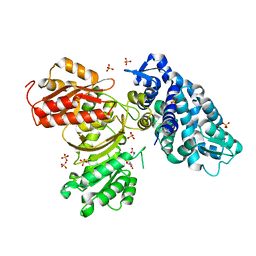 | | AprA Methyltransferase 1 - GNAT in complex with SAH | | Descriptor: | AprA Methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
5UHU
 
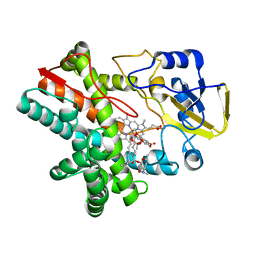 | |
3GWZ
 
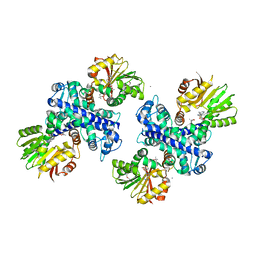 | | Structure of the Mitomycin 7-O-methyltransferase MmcR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MmcR, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
3GXO
 
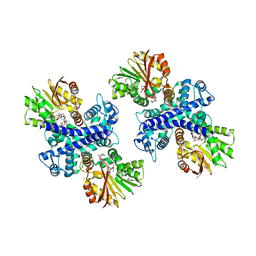 | | Structure of the Mitomycin 7-O-methyltransferase MmcR with bound Mitomycin A | | Descriptor: | CALCIUM ION, MmcR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
6O5G
 
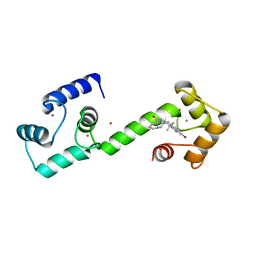 | | Calmodulin in complex with isomalbrancheamide D | | Descriptor: | (5aS,12aS,13aS)-9-bromo-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7 ,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2019-03-02 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3SSO
 
 | |
3SSN
 
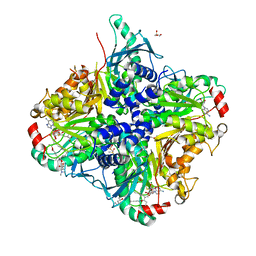 | | MycE Methyltransferase from the Mycinamycin Biosynthetic Pathway in Complex with Mg, SAH, and Mycinamycin VI | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A new structural form in the SAM/metal-dependent o‑methyltransferase family: MycE from the mycinamicin biosynthetic pathway.
J.Mol.Biol., 413, 2011
|
|
3SSM
 
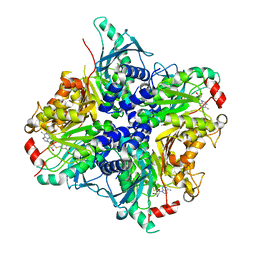 | |
3TO3
 
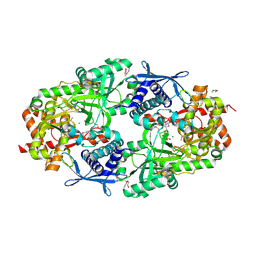 | | Crystal Structure of Petrobactin Biosynthesis Protein AsbB from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kim, Y, Eschenfeldt, W, Stols, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-03 | | Release date: | 2011-10-05 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Functional and Structural Analysis of the Siderophore Synthetase AsbB through Reconstitution of the Petrobactin Biosynthetic Pathway from Bacillus anthracis.
J.Biol.Chem., 287, 2012
|
|
3U1T
 
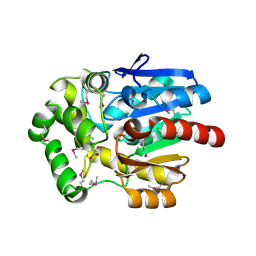 | | Haloalkane Dehalogenase, DmmA, of marine microbial origin | | Descriptor: | CHLORIDE ION, DmmA Haloalkane Dehalogenase, MALONATE ION | | Authors: | Gehret, J.J, Smith, J.L. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of DmmA, a marine haloalkane dehalogenase.
Protein Sci., 21, 2012
|
|
3V7I
 
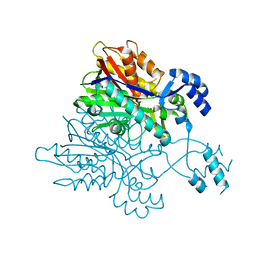 | |
3KG6
 
 | |
3KG8
 
 | |
3KG9
 
 | |
3KG7
 
 | |
5THZ
 
 | |
5THY
 
 | |
5TZ7
 
 | |
5TZ6
 
 | |
5TZ5
 
 | |
5WGT
 
 | | Crystal Structure of MalA' H253A, premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WM4
 
 | | Crystal Structure of CahJ in Complex with 6-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-6-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WM2
 
 | |
