6DLT
 
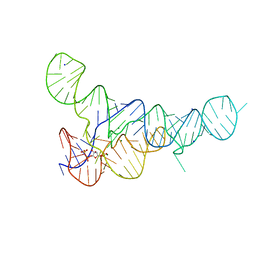 | | PRPP Riboswitch bound to PRPP, native structure | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PRPP Riboswitch, ... | | Authors: | Peselis, A, Serganov, A. | | Deposit date: | 2018-06-02 | | Release date: | 2018-11-14 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands.
Nat. Chem. Biol., 14, 2018
|
|
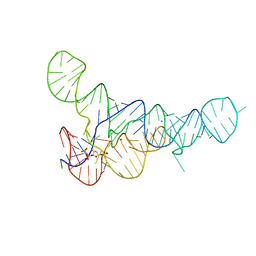
6DLS
 
 | |
6DMC
 
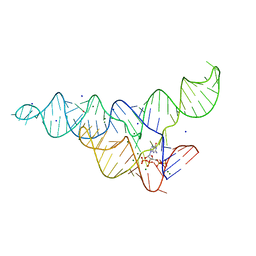 | | ppGpp Riboswitch bound to ppGpp, native structure | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Peselis, A, Serganov, A. | | Deposit date: | 2018-06-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands.
Nat. Chem. Biol., 14, 2018
|
|
6DME
 
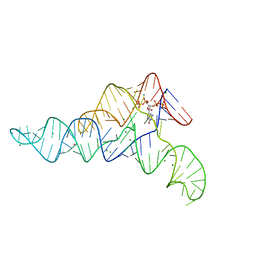 | | ppGpp Riboswitch bound to ppGpp, thallium acetate structure | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Peselis, A, Serganov, A. | | Deposit date: | 2018-06-05 | | Release date: | 2018-11-14 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands.
Nat. Chem. Biol., 14, 2018
|
|
6DLR
 
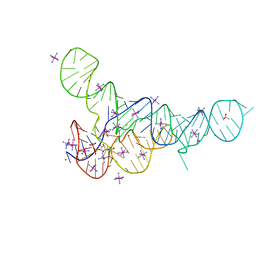 | |
6DMD
 
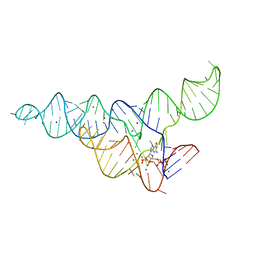 | | ppGpp Riboswitch bound to ppGpp, manganese chloride structure | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Peselis, A, Serganov, A. | | Deposit date: | 2018-06-05 | | Release date: | 2018-11-14 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands.
Nat. Chem. Biol., 14, 2018
|
|
6DNR
 
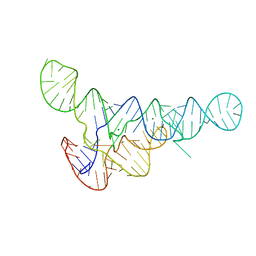 | |
3OWI
 
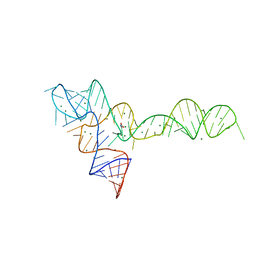 | |
3OXD
 
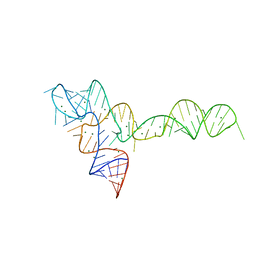 | |
3OWW
 
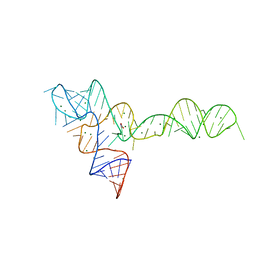 | |
3OXE
 
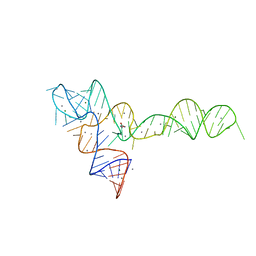 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OWZ
 
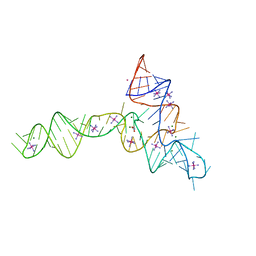 | | Crystal structure of glycine riboswitch, soaked in Iridium | | Descriptor: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXB
 
 | |
3OXM
 
 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXJ
 
 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | Descriptor: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OX0
 
 | |
3SUH
 
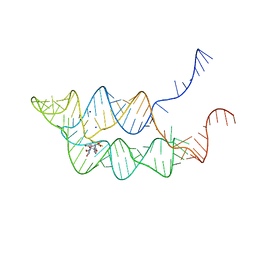 | | Crystal structure of THF riboswitch, bound with 5-formyl-THF | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUX
 
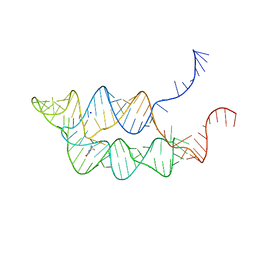 | | Crystal structure of THF riboswitch, bound with THF | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUY
 
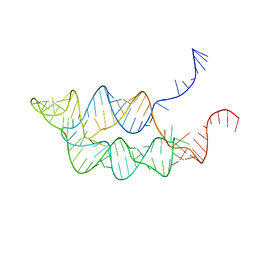 | |
3DW7
 
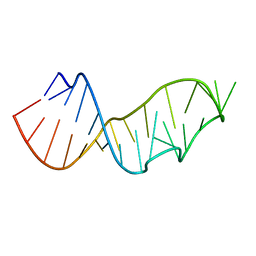 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW5
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW4
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DVZ
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DIM
 
 | |
