7CYX
 
 | |
7FBG
 
 | |
8WEU
 
 | |
8WEV
 
 | |
7XWT
 
 | |
7XWC
 
 | |
7XWV
 
 | |
6L3O
 
 | |
6L3P
 
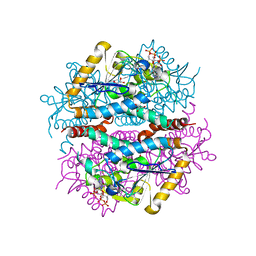 | |
6LKS
 
 | | Effects of zinc ion on oligomerization and pH stability of influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Seok, J, Kim, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Divalent cation-induced conformational changes of influenza virus hemagglutinin.
Sci Rep, 10, 2020
|
|
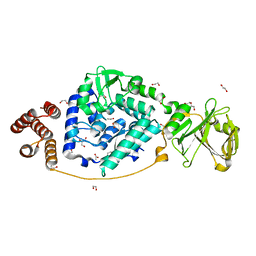
7VGM
 
 | |
7DIB
 
 | |
8JUJ
 
 | |
8JUI
 
 | |
8I4N
 
 | |
8I4Q
 
 | |
6ITK
 
 | |
6ITL
 
 | |
8I4R
 
 | |
