2YVJ
 
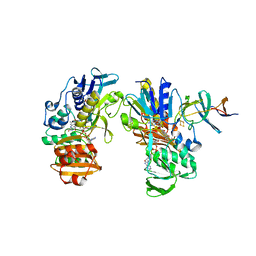 | | Crystal structure of the ferredoxin-ferredoxin reductase (BPHA3-BPHA4)complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2YVF
 
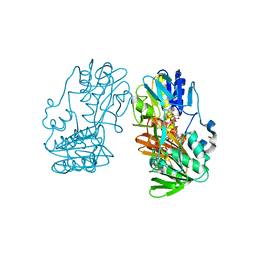 | | Crystal structure of ferredoxin reductase BPHA4 (hydroquinone) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2YVG
 
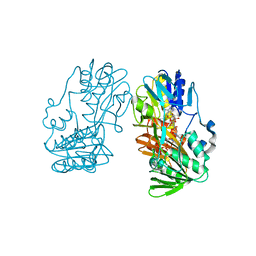 | | crystal structure of ferredoxin reductase, BPHA4 (blue-semiquinone) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
5X94
 
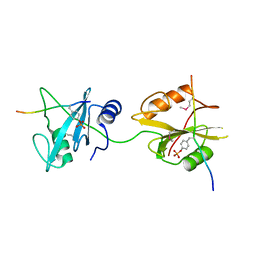 | | Crystal structure of SHP2_SH2-CagA EPIYA_D peptide complex | | Descriptor: | Cag pathogenicity island protein, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2017-03-05 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential Mechanisms for SHP2 Binding and Activation Are Exploited by Geographically Distinct Helicobacter pylori CagA Oncoproteins.
Cell Rep, 20, 2017
|
|
5X7B
 
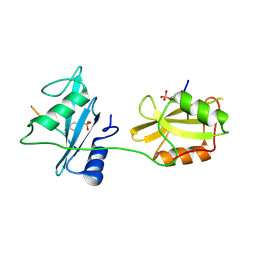 | | Crystal structure of SHP2_SH2-CagA EPIYA_C peptide complex | | Descriptor: | CagA, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2017-02-24 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Differential Mechanisms for SHP2 Binding and Activation Are Exploited by Geographically Distinct Helicobacter pylori CagA Oncoproteins.
Cell Rep, 20, 2017
|
|
7E18
 
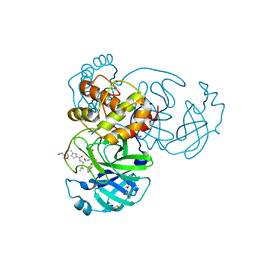 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor YH-53 | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1ab | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7E19
 
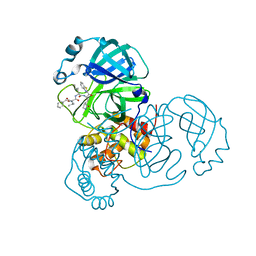 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor SH-5 | | Descriptor: | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7EM6
 
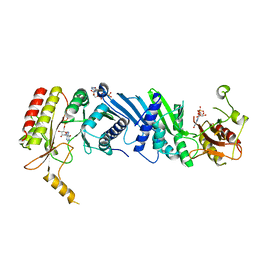 | | Crystal structure of the PI5P4Kbeta N203D-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM2
 
 | | Crystal structure of the PI5P4Kbeta-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM1
 
 | | Crystal structure of the PI5P4Kbeta-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM7
 
 | | Crystal structure of the PI5P4Kbeta N203D-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM5
 
 | | Crystal structure of the PI5P4Kbeta F205L-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM3
 
 | | Crystal structure of the PI5P4Kbeta-2a-ATP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM4
 
 | | Crystal structure of the PI5P4Kbeta F205L-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM8
 
 | | Crystal structure of the PI5P4Kbeta T201M-2a-ATP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7DNM
 
 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DNN
 
 | | Crystal structure of the AgCarB2-C2 complex with homoorientin | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-6-[(2S,3R,4R,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-5,7-bis(oxidanyl)chromen-4-one, AP_endonuc_2 domain-containing protein, AgCarC2, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
7DVE
 
 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D98
 
 | |
6K4H
 
 | | Crystal structure of the PI5P4Kbeta-AMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
8JW3
 
 | | The crystal structure of the viral terpene synthase from Orpheovirus IHUMI-LCC2 | | Descriptor: | SULFATE ION, Terpenoid synthase | | Authors: | Jung, Y, Mitsuhashi, T, Senda, M, Sato, S, Senda, T, Fujita, M. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Function and Structure of a Terpene Synthase Encoded in a Giant Virus Genome.
J.Am.Chem.Soc., 145, 2023
|
|
6ISV
 
 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | Descriptor: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
2E50
 
 | | Crystal structure of SET/TAF-1beta/INHAT | | Descriptor: | Protein SET, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Muto, S, Senda, M, Senda, T, Horikoshi, M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4H4W
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/T176R/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
4H4S
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
