3TD2
 
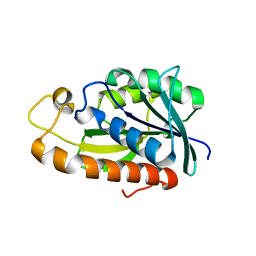 | | Crystal structures of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis - Form 5 | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Ahmad, R, Varshney, U, Vijayan, M. | | Deposit date: | 2011-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of new crystal forms of Mycobacterium tuberculosis peptidyl-tRNA hydrolase and functionally important plasticity of the molecule
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3TCK
 
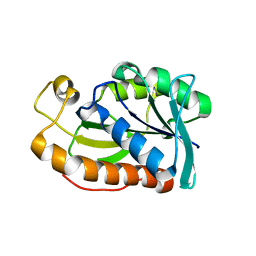 | | Crystal structure of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis - Form 4 | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Ahmad, R, Varshney, U, Vijayan, M. | | Deposit date: | 2011-08-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of new crystal forms of Mycobacterium tuberculosis peptidyl-tRNA hydrolase and functionally important plasticity of the molecule
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3TCN
 
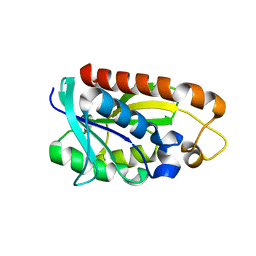 | | Crystal structures of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis - Form 2 grown in presence of Pentaglycine | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Ahmad, R, Varshney, U, Vijayan, M. | | Deposit date: | 2011-08-09 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of new crystal forms of Mycobacterium tuberculosis peptidyl-tRNA hydrolase and functionally important plasticity of the molecule
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3TD6
 
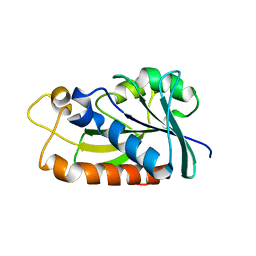 | | Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis from trigonal partially dehydrated crystal | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Ahmad, R, Varshney, U, Vijayan, M. | | Deposit date: | 2011-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of new crystal forms of Mycobacterium tuberculosis peptidyl-tRNA hydrolase and functionally important plasticity of the molecule
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8RFH
 
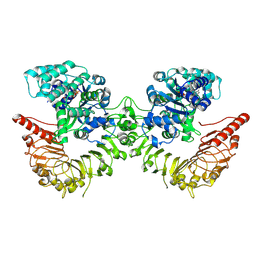 | |
2Z2I
 
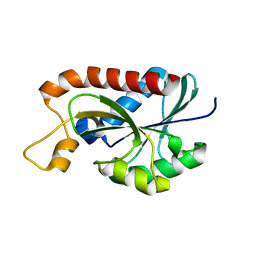 | | Crystal structure of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Roy, S, Singh, N.S, Sangeetha, R, Varshney, U, Vijayan, M. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Plasticity and Enzyme Action: Crystal Structures of Mycobacterium tuberculosis Peptidyl-tRNA Hydrolase
J.Mol.Biol., 372, 2007
|
|
2Z2K
 
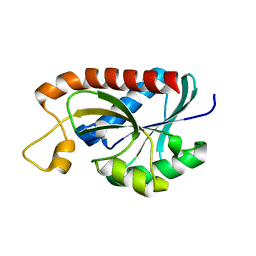 | | Crystal structure of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Roy, S, Singh, N.S, Sangeetha, R, Varshney, U, Vijayan, M. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Plasticity and Enzyme Action: Crystal Structures of Mycobacterium tuberculosis Peptidyl-tRNA Hydrolase
J.Mol.Biol., 372, 2007
|
|
2Z2J
 
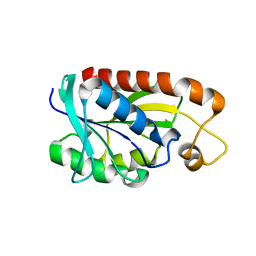 | | Crystal structure of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Roy, S, Singh, N.S, Sangeetha, R, Varshney, U, Vijayan, M. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Plasticity and Enzyme Action: Crystal Structures of Mycobacterium tuberculosis Peptidyl-tRNA Hydrolase
J.Mol.Biol., 372, 2007
|
|
4KB4
 
 | | Crystal structure of ribosome recycling factor mutant R31A from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KDD
 
 | | Structure of Mycobacterium tuberculosis ribosome recycling factor in presence of detergent | | Descriptor: | CADMIUM ION, DECYL-BETA-D-MALTOPYRANOSIDE, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-24 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KAW
 
 | | Crystal structure of ribosome recycling factor mutant R39G from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KB2
 
 | | Crystal structure of ribosome recycling factor mutant R109A from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
4KC6
 
 | | Crystal structure of C-terminal deletion mutant of ribosome recycling factor from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome-recycling factor | | Authors: | Selvaraj, M, Govindan, A, Seshadri, A, Dubey, B, Varshney, U, Vijayan, M. | | Deposit date: | 2013-04-24 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants.
J.Biosci., 38, 2013
|
|
7ZTD
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 3.2, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
7ZTC
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 1.6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 1.6, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
6G0Y
 
 | | X-ray structure of M-21 protein complex | | Descriptor: | Matrix M2-1, Phosphoprotein, ZINC ION | | Authors: | Edwards, T.A, Barr, J. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structure of the Human Respiratory Syncytial Virus M2-1 Protein Bound to the Interaction Domain of the Phosphoprotein P Defines the Orientation of the Complex.
Mbio, 9, 2018
|
|
8BBO
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-36 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGH@ protein, Immunoglobulin kappa light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BBN
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-10 and EY6A Fabs | | Descriptor: | BA.2-10 heavy chain, BA.2-10 light chain, EY6A Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8R1C
 
 | | SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-2 fab heavy chain, SD1-2 fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R1D
 
 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8BV0
 
 | | Binary complex between the NB-ARC domain from the Tomato immune receptor NRC1 and the SPRY domain-containing effector SS15 from the potato cyst nematode | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NRC1, Truncated secreted SPRY domain-containing protein 15 (Fragment) | | Authors: | Contreras, M.P, Pai, H, Muniyandi, S, Toghani, A, Lawson, D.M, Tumtas, Y, Duggan, C, Yuen, E.L.H, Stevenson, C.E.M, Harant, A, Wu, C.H, Bozkurt, T.O, Kamoun, S, Derevnina, L. | | Deposit date: | 2022-12-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Sci Adv, 9, 2023
|
|
8QZR
 
 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
6XXE
 
 | |
6XXD
 
 | |
