2OJY
 
 | |
6FP0
 
 | |
6FOX
 
 | |
6GW3
 
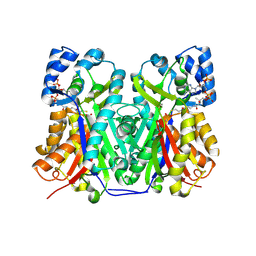 | | Structure of TKS from Cannabis sativa in complex with CoA | | Descriptor: | 3,5,7-trioxododecanoyl-CoA synthase, 3,5,7-trioxododecanoyl-CoA synthase,3,5,7-trioxododecanoyl-CoA synthase, COENZYME A, ... | | Authors: | Karuppiah, V, Leys, D. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of a PKS class III from Cannabis sativa
To Be Published
|
|
6GIA
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant I107A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI7
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant L25I | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI8
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant L25A | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6GI9
 
 | | Crystal structure of pentaerythritol tetranitrate reductase (PETNR) mutant I107L | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Levy, C.W. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Nonequivalence of Second Sphere "Noncatalytic" Residues in Pentaerythritol Tetranitrate Reductase in Relation to Local Dynamics Linked to H-Transfer in Reactions with NADH and NADPH Coenzymes.
Acs Catalysis, 8, 2018
|
|
6I3C
 
 | |
6I3D
 
 | |
3CLR
 
 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
3CLU
 
 | | Crystal structure of the R236K mutant from Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
2HKR
 
 | |
3CLT
 
 | | Crystal structure of the R236E mutant of Methylophilus methylotrophus ETF | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
2HJ4
 
 | |
3CLS
 
 | | Crystal structure of the R236C mutant of ETF from Methylophilus methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
2HJB
 
 | |
2HKM
 
 | |
2HXC
 
 | |
2I0R
 
 | |
2I0S
 
 | |
2I0T
 
 | |
1H61
 
 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H63
 
 | | Structure of the reduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1GWJ
 
 | | Morphinone reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, MORPHINONE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2002-03-18 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bacterial Morphinone Reductase and Properties of the C191A Mutant Enzyme.
J.Biol.Chem., 277, 2002
|
|
