7QEY
 
 | | human Connexin 26 class 1 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEW
 
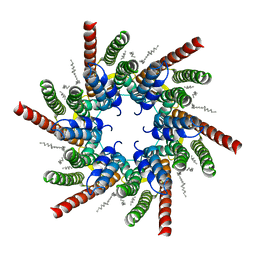 | | human Connexin 26 class 2 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEQ
 
 | | human Connexin 26 dodecamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEU
 
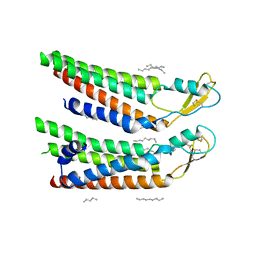 | | human Connexin 26 at 55mmHg PCO2, pH7.4: two masked subunits, class B | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEO
 
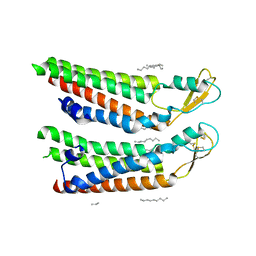 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEV
 
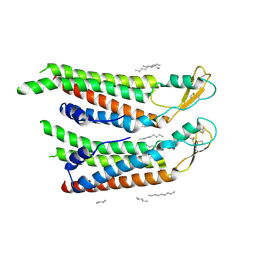 | | human Connexin 26 at 55mm Hg PCO2, pH7.4:two masked subunits, class D | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QES
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class A | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
2NVO
 
 | |
3ONR
 
 | | Crystal structure of the calcium chelating immunodominant antigen, calcium dodecin (Rv0379),from Mycobacterium tuberculosis with a novel calcium-binding site | | Descriptor: | CALCIUM ION, FORMIC ACID, PLATINUM (II) ION, ... | | Authors: | Arulandu, A, Sacchettini, J.C. | | Deposit date: | 2010-08-30 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of calcium dodecin (Rv0379), from Mycobacterium tuberculosis with a unique calcium-binding site.
Protein Sci., 20, 2011
|
|
2YGT
 
 | | Clostridium perfringens delta-toxin | | Descriptor: | DELTA TOXIN, GLYCEROL, IMIDAZOLE, ... | | Authors: | Huyet, J, Naylor, C.E, Gibert, M, Popoff, M.R, Basak, A.K. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Clostridium Perfringens Delta Toxin Pore Formation.
Plos One, 8, 2013
|
|
4P5H
 
 | |
6S1M
 
 | | Human polymerase delta holoenzyme Conformer 1 | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6S1N
 
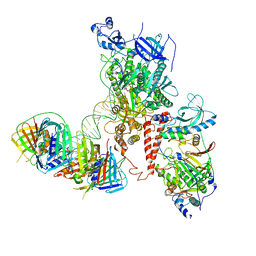 | | Human polymerase delta holoenzyme Conformer 2 | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6S1O
 
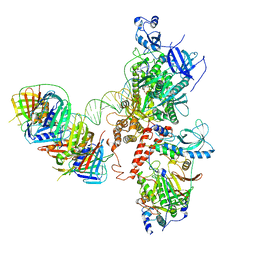 | | Human polymerase delta holoenzyme Conformer 3 | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6TNZ
 
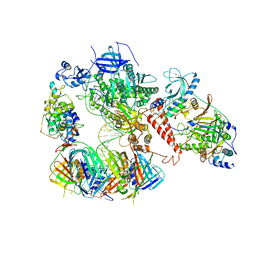 | | Human polymerase delta-FEN1-PCNA toolbelt | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6TNY
 
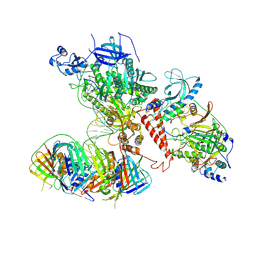 | | Processive human polymerase delta holoenzyme | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-12-10 | | Release date: | 2019-12-18 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
5LJW
 
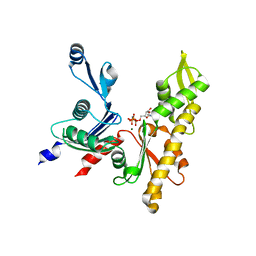 | | MamK non-polymerising A278D mutant bound to AMPPNP | | Descriptor: | Actin-like ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lowe, J. | | Deposit date: | 2016-07-20 | | Release date: | 2016-11-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LJV
 
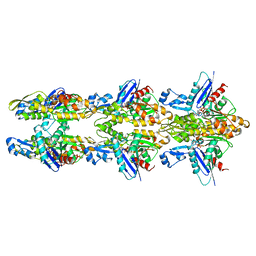 | | MamK double helical filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like ATPase, MAGNESIUM ION | | Authors: | Lowe, J. | | Deposit date: | 2016-07-21 | | Release date: | 2016-11-16 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5MPS
 
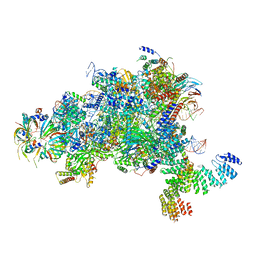 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5GAO
 
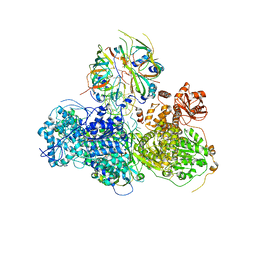 | | Head region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2, Saccharomyces cerevisiae strain UOA_M2 chromosome 5 sequence, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5GAM
 
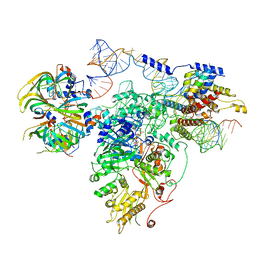 | | Foot region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-splicing factor 8, Pre-mRNA-splicing factor SNU114, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-02-03 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Angstrom resolution
Nature, 530, 2016
|
|
5GAN
 
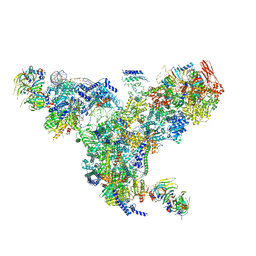 | | The overall structure of the yeast spliceosomal U4/U6.U5 tri-snRNP at 3.7 Angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-processing factor 31, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
