1H88
 
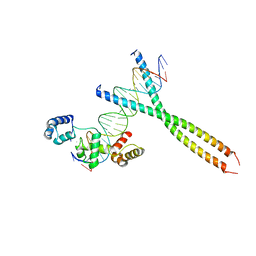 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | 分子名称: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-29 | | 公開日 | 2002-01-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
5BRO
 
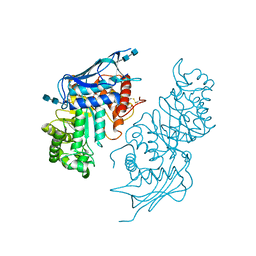 | | Crystal structure of modified HexB (modB) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase subunit beta, FORMIC ACID, ... | | 著者 | Kitakaze, K, Maita, N, Itoh, K. | | 登録日 | 2015-06-01 | | 公開日 | 2016-05-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protease-resistant modified human beta-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model.
J.Clin.Invest., 126, 2016
|
|
4EPS
 
 | |
8JYN
 
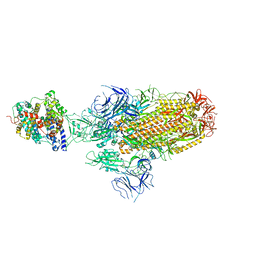 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYM
 
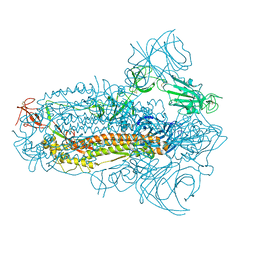 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYK
 
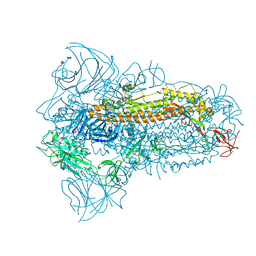 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
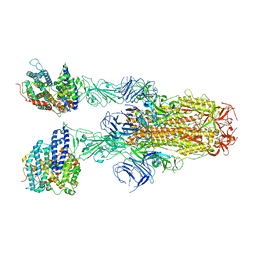 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYP
 
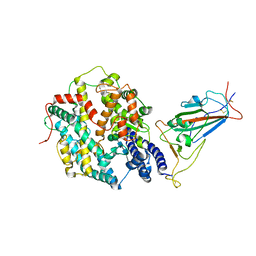 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
1TCJ
 
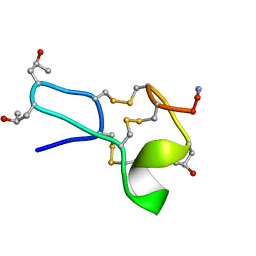 | |
1TCG
 
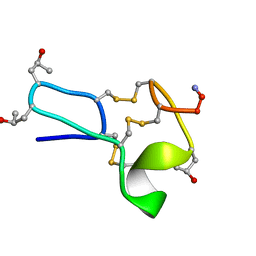 | |
4H40
 
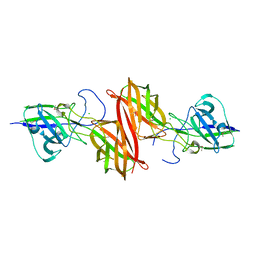 | |
4GPV
 
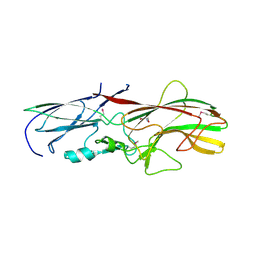 | |
8IOT
 
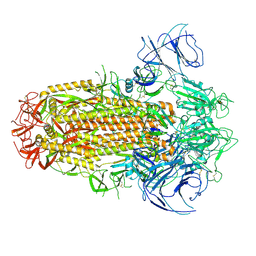 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
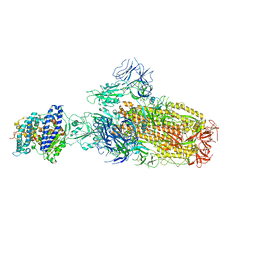 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOS
 
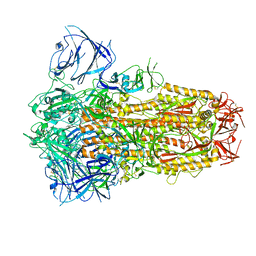 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IF2
 
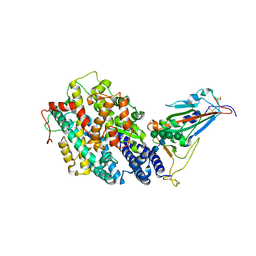 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Kimura, K, Suzuki, T, Hashiguchi, T. | | 登録日 | 2023-02-17 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-05-24 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
8IOV
 
 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
4YBG
 
 | | Crystal structure of the MAEL domain of Drosophila melanogaster Maelstrom | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Protein maelstrom, ... | | 著者 | Matsumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | 登録日 | 2015-02-18 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Crystal Structure and Activity of the Endoribonuclease Domain of the piRNA Pathway Factor Maelstrom
Cell Rep, 11, 2015
|
|
1HJB
 
 | |
1HJC
 
 | |
1IO4
 
 | |
1TCH
 
 | |
1TCK
 
 | |
8A51
 
 | | Crystal structure of HSF2BP-BRME1 complex | | 分子名称: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | 著者 | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | 登録日 | 2022-06-13 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
8A50
 
 | | Crystal structure of HSF2BP-ALPHA1 tetramer | | 分子名称: | Heat shock factor 2-binding protein, PHOSPHATE ION | | 著者 | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | 登録日 | 2022-06-13 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.484 Å) | | 主引用文献 | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
