1RE2
 
 | | HUMAN LYSOZYME LABELLED WITH TWO 2',3'-EPOXYPROPYL BETA-GLYCOSIDE OF N-ACETYLLACTOSAMINE | | Descriptor: | GLYCEROL, PROTEIN (LYSOZYME), beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1998-11-05 | | Release date: | 1999-05-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual affinity labeling of the active site of human lysozyme with an N-acetyllactosamine derivative: first ligand assisted recognition of the second ligand.
Biochemistry, 38, 1999
|
|
1REM
 
 | | HUMAN LYSOZYME WITH MAN-B1,4-GLCNAC COVALENTLY ATTACHED TO ASP53 | | Descriptor: | LYSOZYME, NITRATE ION, R-1,2-PROPANEDIOL, ... | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1998-01-14 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of human lysozyme labelled with 2',3'-epoxypropyl beta-glycoside of man-beta1,4-GlcNAc. Structural change and recognition specificity at subsite B.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3FFD
 
 | | Structure of parathyroid hormone-related protein complexed to a neutralizing monoclonal antibody | | Descriptor: | Monoclonal antibody, heavy chain, Fab fragment, ... | | Authors: | Mckinstry, W.J, Polekhina, G, Diefenbach-Jagger, H, Ho, P.W.M, Sato, K, Onuma, E, Gillespie, M.T, Martin, T.J, Parker, M.W. | | Deposit date: | 2008-12-03 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody discrimination between two hormones that recognize the parathyroid hormone receptor
J.Biol.Chem., 284, 2009
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
1NC8
 
 | | HIGH-RESOLUTION SOLUTION NMR STRUCTURE OF THE MINIMAL ACTIVE DOMAIN OF THE HUMAN IMMUNODEFICIENCY VIRUS TYPE-2 NUCLEOCAPSID PROTEIN, 15 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Kodera, Y, Sato, K, Tsukahara, T, Komatsu, H, Maeda, T, Kohno, T. | | Deposit date: | 1998-05-14 | | Release date: | 1999-05-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein.
Biochemistry, 37, 1998
|
|
1OAV
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1OAW
 
 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1V4Q
 
 | |
8JUF
 
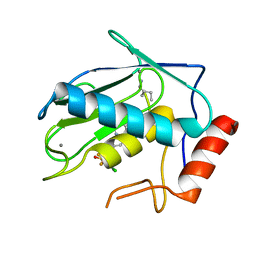 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8JUD
 
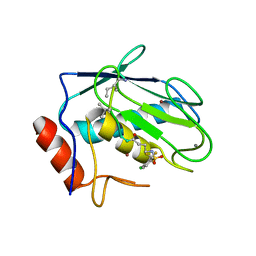 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8JUG
 
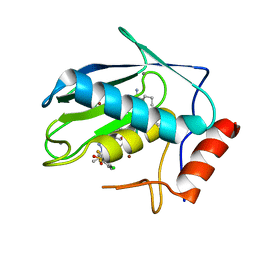 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
6KJK
 
 | | Structure of the N-terminal domain of PorA | | Descriptor: | N-terminal domain of PorA (28-171) | | Authors: | Handa, Y, Sato, K, Shoji, M, Nakayama, K, Imada, K. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-22 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PorA, a conserved C-terminal domain-containing protein, impacts the PorXY-SigP signaling of the type IX secretion system.
Sci Rep, 10, 2020
|
|
1F3K
 
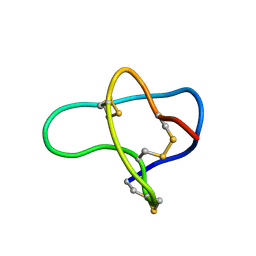 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF OMEGA-CONOTOXIN TXVII, AN L-TYPE CALCIUM CHANNEL BLOCKER | | Descriptor: | OMEGA-CONOTOXIN TXVII | | Authors: | Kobayashi, K, Sasaki, T, Sato, K, Kohno, T. | | Deposit date: | 2000-06-05 | | Release date: | 2000-12-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of omega-conotoxin TxVII, an L-type calcium channel blocker.
Biochemistry, 39, 2000
|
|
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | Descriptor: | IMPERATOXIN A | | Authors: | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | Deposit date: | 2001-04-07 | | Release date: | 2003-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
6K4J
 
 | | Crystal Structure of the the CD9 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | Authors: | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | Deposit date: | 2019-05-24 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
1FU3
 
 | |
1WQB
 
 | |
2A70
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A71
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), orthorhombic crystal form | | Descriptor: | Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6W
 
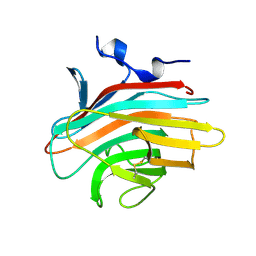 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), metal-free form | | Descriptor: | Emp46p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Y
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), tetragonal crystal form | | Descriptor: | Emp47p (form1), SULFATE ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6V
 
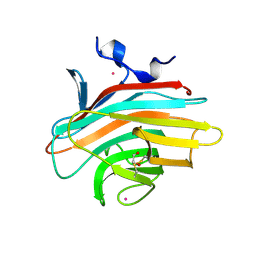 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), potassium-bound form | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Z
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 1 | | Descriptor: | Emp47p (form2) | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
