4KX6
 
 | | Plasticity of the quinone-binding site of the complex II homolog quinol:fumarate reductase | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Singh, P.K, Sarwar, M, Maklashina, E, Kotlyar, V, Rajagukguk, S, Tomasiak, T.M, Cecchini, G, Iverson, T.M. | | 登録日 | 2013-05-24 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Plasticity of the Quinone-binding Site of the Complex II Homolog Quinol:Fumarate Reductase.
J.Biol.Chem., 288, 2013
|
|
4N0E
 
 | | Crystal structure of the K345L variant of the Gi alpha1 subunit bound to GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, SULFATE ION | | 著者 | Thaker, T.M, Preininger, A.M, Sarwar, M, Hamm, H.E, Iverson, T.M. | | 登録日 | 2013-10-01 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Transient Interaction between the Phosphate Binding Loop and Switch I Contributes to the Allosteric Network between Receptor and Nucleotide in G alpha i1.
J.Biol.Chem., 289, 2014
|
|
4N0D
 
 | | Crystal structure of the K345L variant of the Gi alpha1 subunit bound to GTPgammaS | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION, ... | | 著者 | Thaker, T.M, Preininger, A.M, Sarwar, M, Hamm, H.E, Iverson, T.M. | | 登録日 | 2013-10-01 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Transient Interaction between the Phosphate Binding Loop and Switch I Contributes to the Allosteric Network between Receptor and Nucleotide in G alpha i1.
J.Biol.Chem., 289, 2014
|
|
3P4P
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with fumarate | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Tomasiak, T.M, Archuleta, T.L, Andr ll, J, Luna-Ch vez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | 登録日 | 2010-10-06 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4Q
 
 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | 登録日 | 2010-10-06 | | 公開日 | 2010-12-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4R
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with glutarate | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | 登録日 | 2010-10-07 | | 公開日 | 2010-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4S
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with a 3-nitropropionate adduct | | 分子名称: | 3-NITROPROPANOIC ACID, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | 著者 | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | 登録日 | 2010-10-07 | | 公開日 | 2010-12-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
4XAC
 
 | | Crystal Structure of EvdO2 from Micromonospora carbonacea var. aurantiaca complexed with 2-oxoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, EvdO2, IMIDAZOLE, ... | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-13 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XCA
 
 | | Crystal Structure of HygX from Streptomyces hygroscopicus with nickel and 2-oxoglutarate bound | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-OXOGLUTARIC ACID, CESIUM ION, ... | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XAA
 
 | | Crystal Structure of AviO1 from Streptomyces viridochromogenes Tue57 | | 分子名称: | NICKEL (II) ION, Putative oxygenase | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-13 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XAB
 
 | | Crystal Structure of EvdO2 from Micromonospora carbonacea var. aurantiaca | | 分子名称: | EvdO2, IMIDAZOLE, NICKEL (II) ION | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-13 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XCB
 
 | | Crystal Structure of HygX from Streptomyces hygroscopicus with nickel, 2-oxoglutarate, and hygromycin B bound | | 分子名称: | 2-OXOGLUTARIC ACID, HYGROMYCIN B VARIANT, NICKEL (II) ION, ... | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XBZ
 
 | | Crystal Structure of EvdO1 from Micromonospora carbonacea var. aurantiaca | | 分子名称: | EvdO1, GLYCEROL, NICKEL (II) ION | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XC9
 
 | | Crystal Structure of apo HygX from Streptomyces hygroscopicus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, oxidase/hydroxylase | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZPI
 
 | | Crystal Structure of HygX from Streptomyces hygroscopicus with iron bound | | 分子名称: | FE (II) ION, Putative oxidase/hydroxylase, SUCCINIC ACID | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2015-05-07 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.504 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1B4E
 
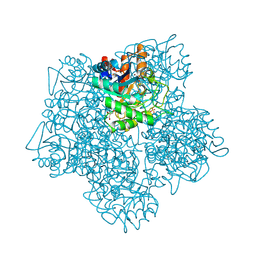 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | 分子名称: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | 著者 | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | 登録日 | 1998-12-19 | | 公開日 | 1999-12-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
1AW5
 
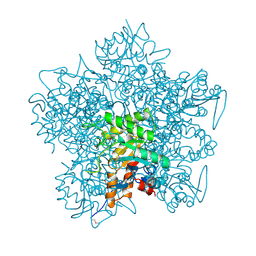 | |
5HMS
 
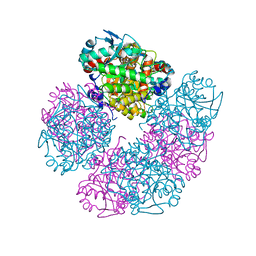 | | X-ray structure of human recombinant 5-aminolaevulinic acid dehydratase (hrALAD). | | 分子名称: | Delta-aminolevulinic acid dehydratase, ZINC ION | | 著者 | Butler, D, Erskine, P.T, Cooper, J.B, Shoolingin-Jordan, P.M. | | 登録日 | 2016-01-17 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5HNR
 
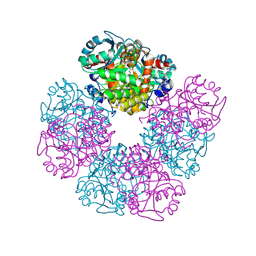 | | The X-ray structure of octameric human native 5-aminolaevulinic acid dehydratase. | | 分子名称: | DELTA-AMINO VALERIC ACID, Delta-aminolevulinic acid dehydratase, SULFATE ION, ... | | 著者 | Mills-Davies, N.L, Thompson, D, Shoolingin-Jordan, P.M, Erskine, P.T, Cooper, J.B. | | 登録日 | 2016-01-18 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LZL
 
 | | Pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase | | 分子名称: | Delta-aminolevulinic acid dehydratase, ZINC ION | | 著者 | Azim, N, Erskine, P.T, Guo, J, Cooper, J.B. | | 登録日 | 2016-09-30 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MHB
 
 | | Product-Complex of E.coli 5-Amino Laevulinic Acid Dehydratase | | 分子名称: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, Delta-aminolevulinic acid dehydratase, GLYCEROL, ... | | 著者 | Norton, E, Erskine, P.T, Shoolingin-Jordan, P.M, Cooper, J.B. | | 登録日 | 2016-11-23 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3EQ1
 
 | | The Crystal Structure of Human Porphobilinogen Deaminase at 2.8A resolution | | 分子名称: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase, SULFATE ION | | 著者 | Kolstoe, S.E, Gill, R, Mohammed, F, Wood, S.P. | | 登録日 | 2008-09-30 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of human porphobilinogen deaminase at 2.8 A: the molecular basis of acute intermittent porphyria
Biochem.J., 420, 2009
|
|
