1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1TUO
 
 | | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8 | | Descriptor: | Putative phosphomannomutase | | Authors: | Misaki, S, Suzuki, S, Fujimoto, S, Sakurai, M, Kobayashi, M, Nishijima, K, Kunishima, N, Sugawara, M, Kuroishi, C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1WPW
 
 | | Crystal Structure of IPMDH from Sulfolobus tokodaii | | Descriptor: | 3-isopropylmalate dehydrogenase, MAGNESIUM ION | | Authors: | Hirose, R, Sakurai, M, Suzuki, T, Moriyama, H, Sato, T, Yamagishi, A, Oshima, T, Tanaka, N. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of IPMDH from Sulfolobus tokodaii
To be Published
|
|
5AXH
 
 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
5AXG
 
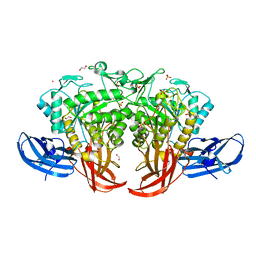 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Dextranase, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
7DC8
 
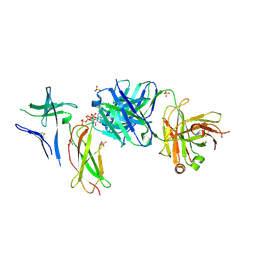 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7DC7
 
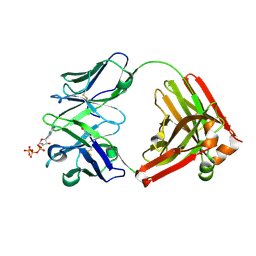 | | Crystal structure of D12 Fab-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | Authors: | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
9C6M
 
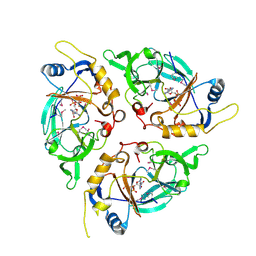 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase, K73M mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA ligase1 | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9C6L
 
 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, RNA ligase1, ... | | Authors: | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-02 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7DRU
 
 | |
3NJY
 
 | | Crystal structure of JMJD2A complexed with 5-carboxy-8-hydroxyquinoline | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | King, O.N.F, Clifton, I.J, Wang, M, Maloney, D.J, Jadhav, A, Oppermann, U, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Plos One, 5, 2010
|
|
5Z7M
 
 | | SmChiA sliding-intermediate with chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase A, ... | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
5Z7N
 
 | | SmChiA sliding-intermediate with chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase A, ... | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
5Z7O
 
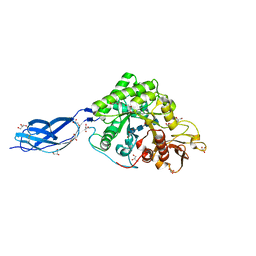 | | SmChiA sliding-intermediate with chitotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, GLYCEROL | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
5Z7P
 
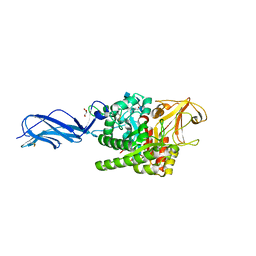 | | SmChiA sliding-intermediate with chitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, GLYCEROL | | Authors: | Nakamura, A, Iino, R. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin.
Nat Commun, 9, 2018
|
|
1XAC
 
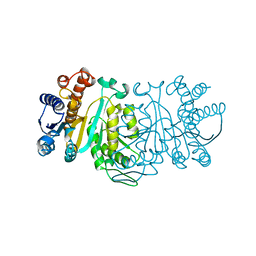 | |
1XAD
 
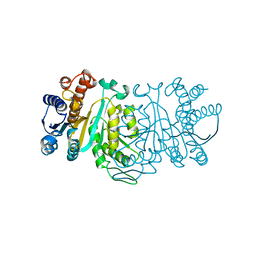 | |
