1SAF
 
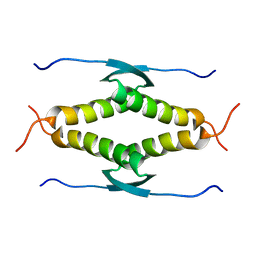 | |
1J02
 
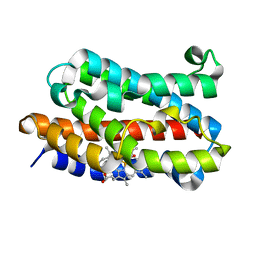 | | Crystal Structure of Rat Heme Oxygenase-1-Heme Bound to NO | | Descriptor: | HEME OXYGENASE 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
2ZVU
 
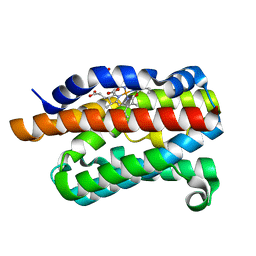 | | Crystal structure of rat heme oxygenase-1 in complex with ferrous verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, FORMIC ACID, Heme oxygenase 1 | | Authors: | Sato, H, Sugishima, M, Fukuyama, K, Noguchi, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side
Biochem.J., 419, 2009
|
|
3A4P
 
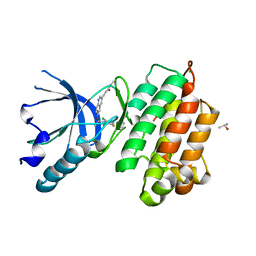 | | human c-MET kinase domain complexed with 6-benzyloxyquinoline inhibitor | | Descriptor: | (2E)-3-{6-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]quinolin-3-yl}-N-methylprop-2-enamide, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Fukami, T.A, Kadono, S, Yamamuro, M, Matsuura, T. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of 6-benzyloxyquinolines as c-Met selective kinase inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6K3D
 
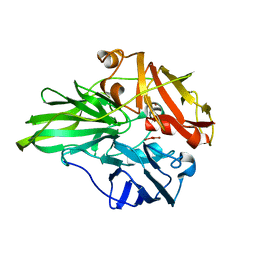 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
2DY5
 
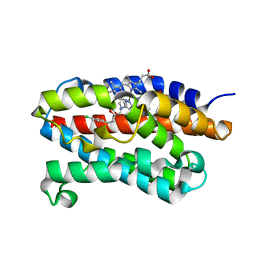 | | Crystal structure of rat heme oxygenase-1 in complex with heme and 2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-1,3-dioxolane | | Descriptor: | 1-({2-[2-(4-CHLOROPHENYL)ETHYL]-1,3-DIOXOLAN-2-YL}METHYL)-1H-IMIDAZOLE, CHLORIDE ION, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Takahashi, H, Fukuyama, K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic and biochemical characterization of the inhibitory action of an imidazole-dioxolane compound on heme oxygenase
Biochemistry, 46, 2007
|
|
1SAK
 
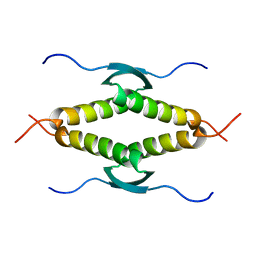 | |
1SAL
 
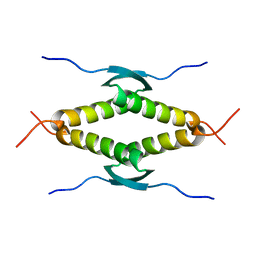 | |
1SAE
 
 | |
1Q80
 
 | | Solution structure and dynamics of Nereis sarcoplasmic calcium binding protein | | Descriptor: | Sarcoplasmic calcium-binding protein | | Authors: | Rabah, G, Popescu, R, Cox, J.A, Engelborghs, Y, Craescu, C.T. | | Deposit date: | 2003-08-20 | | Release date: | 2004-09-21 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure and internal dynamics of NSCP, a compact calcium-binding protein.
Febs J., 272, 2005
|
|
2CMK
 
 | | CYTIDINE MONOPHOSPHATE KINASE IN COMPLEX WITH CYTIDINE-DI-PHOSPHATE | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PROTEIN (CYTIDINE MONOPHOSPHATE KINASE), SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Briozzo, P. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
Structure, 6, 1998
|
|
2E7E
 
 | |
3W0T
 
 | | Human Glyoxalase I with an N-hydroxypyridone derivative inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lactoylglutathione lyase, N-[3-(1-hydroxy-6-oxo-4-phenyl-1,6-dihydropyridin-2-yl)phenyl]methanesulfonamide, ... | | Authors: | Fukami, T.A, Irie, M, Matsuura, T. | | Deposit date: | 2012-11-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | N-Hydroxypyridone-based glyoxalase I inhibitors mimicking binding interactions of the substrate
To be Published
|
|
3VW9
 
 | | Human Glyoxalase I with an N-hydroxypyridone inhibitor | | Descriptor: | 1-hydroxy-6-[1-(3-methoxypropyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-4-phenylpyridin-2(1H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lactoylglutathione lyase, ... | | Authors: | Fukami, T.A, Irie, M, Matsuura, T. | | Deposit date: | 2012-08-10 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Design and evaluation of azaindole-substituted N-hydroxypyridones as glyoxalase I inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3W0U
 
 | | human Glyoxalase I with an N-hydroxypyridone inhibitor | | Descriptor: | Lactoylglutathione lyase, N-[3-(1-Hydroxy-6-oxo-4-phenyl-1,6-dihydro-pyridin-2-yl)-5-methanesulfonylamino-phenyl]-methanesulfonamide, ZINC ION | | Authors: | Fukami, T.A, Irie, M, Matsuura, T. | | Deposit date: | 2012-11-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | N-Hydroxypyridone-based glyoxalase I inhibitors mimicking binding interactions of the substrate
to be published
|
|
3SAK
 
 | |
