1DJU
 
 | | CRYSTAL STRUCTURE OF AROMATIC AMINOTRANSFERASE FROM PYROCOCCUS HORIKOSHII OT3 | | Descriptor: | AROMATIC AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsui, I, Matsui, E, Sakai, Y, Kikuchi, H, Kawarabayashi, H. | | Deposit date: | 1999-12-06 | | Release date: | 2001-04-11 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii.
J.Biol.Chem., 275, 2000
|
|
1IRY
 
 | | Solution structure of the hMTH1, a nucleotide pool sanitization enzyme | | Descriptor: | hMTH1 | | Authors: | Mishima, M, Itoh, N, Sakai, Y, Kamiya, H, Nakabeppu, Y, Shirakawa, M. | | Deposit date: | 2001-10-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates
J.Biol.Chem., 279, 2004
|
|
3AJX
 
 | | Crystal Structure of 3-Hexulose-6-Phosphate Synthase | | Descriptor: | 3-hexulose-6-phosphate synthase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kita, A, Orita, I, Yurimoto, H, Kato, N, Sakai, Y, Miki, K. | | Deposit date: | 2010-06-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of 3-hexulose-6-phosphate synthase, a member of the orotidine 5'-monophosphate decarboxylase suprafamily
Proteins, 78, 2010
|
|
8JJE
 
 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3UG0
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the simplified genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3MYU
 
 | | Mycoplasma genitalium MG289 | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ACETATE ION, High affinity transport system protein p37 | | Authors: | Sippel, K.H, Boehlein, S.K, Govindasamy, L, Namiki, K, Satai, Y, Quirit, J.G, Agbandje-McKenna, M, Goodison, S, Rosser, C.J, Sankaran, B, McKenna, R. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into Mycoplasma genitalium metabolism revealed by the structure of MG289, an extracytoplasmic thiamine binding lipoprotein.
Proteins, 79, 2011
|
|
7DMU
 
 | |
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1GDE
 
 | | CRYSTAL STRUCTURE OF PYROCOCCUS PROTEIN A-1 E-FORM | | Descriptor: | ASPARTATE AMINOTRANSFERASE, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-23 | | Release date: | 2001-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
3UX1
 
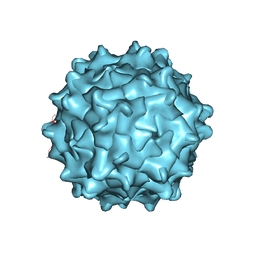 | | Structural Characterization of Adeno-Associated Virus Serotype 9 | | Descriptor: | Capsid protein VP1 | | Authors: | DiMattia, M.A, Nam, H.-J, Van Vliet, K, Mitchell, M, McCall, A, Bennett, A, Gurda, B, McKenna, R, Potter, M, Sakai, Y, Byrne, B.J, Muzyczka, N, Aslanidi, G, Zolotukhin, S, Olson, N, Sinkovitis, R, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the unique properties of adeno-associated virus serotype 9.
J.Virol., 86, 2012
|
|
