7RC4
 
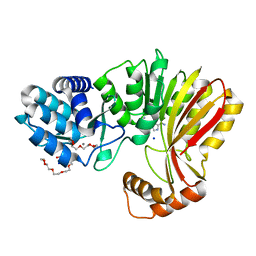 | | Aeronamide N-methyltransferase, AerE (D141A) | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Methyltransferase family protein, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RI3
 
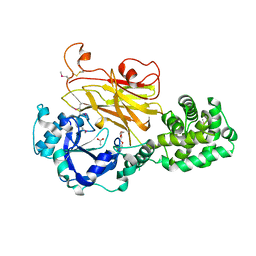 | | Crystal structure of Albireti Toxin, a diphtheria toxin homolog, from Streptomyces albireticuli | | Descriptor: | ACETATE ION, CALCIUM ION, Diphtheria_T domain-containing protein, ... | | Authors: | Sugiman-Marangos, S.N, Gill, S.K, Melnyk, R.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-20 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of distant diphtheria toxin homologs reveal functional determinants of an evolutionarily conserved toxin scaffold.
Commun Biol, 5, 2022
|
|
7RB4
 
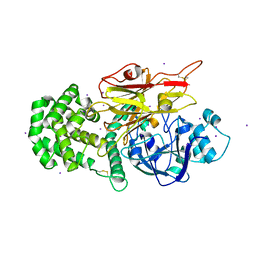 | | Crystal structure of Peptono Toxin, a diphtheria toxin homolog, from Seinonella peptonophila | | Descriptor: | Diphtheria toxin, C domain, IODIDE ION | | Authors: | Sugiman-Marangos, S.N, Gill, S.K, Melnyk, R.A. | | Deposit date: | 2021-07-05 | | Release date: | 2022-04-20 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of distant diphtheria toxin homologs reveal functional determinants of an evolutionarily conserved toxin scaffold.
Commun Biol, 5, 2022
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
8EJY
 
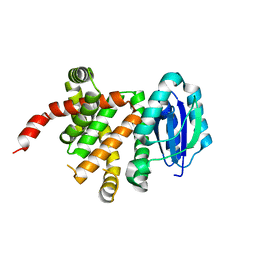 | | [4+2] Aza-Cyclase F293A variant | | Descriptor: | PbtD | | Authors: | Nair, S.K. | | Deposit date: | 2022-09-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Enzymatic Pyridine Aromatization during Thiopeptide Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8EJZ
 
 | | [4+2] Aza-Cyclase Y293F variant | | Descriptor: | PbtD | | Authors: | Nair, S.K. | | Deposit date: | 2022-09-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Pyridine Aromatization during Thiopeptide Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7SOM
 
 | | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | Cilia- and flagella-associated protein 20, FAP147, FAP178, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SJ4
 
 | | Human Trio residues 1284-1959 in complex with Rac1 | | Descriptor: | Ras-related C3 botulinum toxin substrate 1, Triple functional domain protein | | Authors: | Chen, C.-L, Ravala, S.K, Bandekar, S.J, Cash, J, Tesmer, J.J.G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural/functional studies of Trio provide insights into its configuration and show that conserved linker elements enhance its activity for Rac1.
J.Biol.Chem., 298, 2022
|
|
8EW6
 
 | | Anti-human CD8 VHH complex with CD8 alpha | | Descriptor: | Anti-CD8 alpha VHH, T-cell surface glycoprotein CD8 alpha chain | | Authors: | Kiefer, J.R, Williams, S, Davies, C.W, Koerber, J.T, Sriraman, S.K, Yin, Y.P. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Development of an 18 F-labeled anti-human CD8 VHH for same-day immunoPET imaging.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
7SOL
 
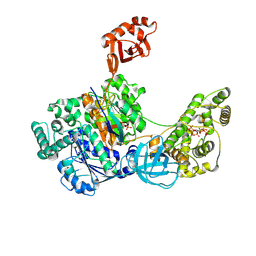 | | Crystal Structures of the bispecific ubiquitin/FAT10 activating enzyme, Uba6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Ubiquitin, ... | | Authors: | Olsen, S.K, Gao, F, Lv, Z. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25000644 Å) | | Cite: | Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Nat Commun, 13, 2022
|
|
8FAZ
 
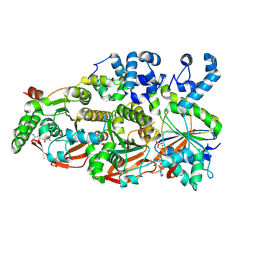 | | Cryo-EM structure of the human BCDX2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8F72
 
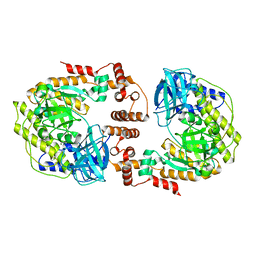 | | Phage P32 gp64- RNA polymerase | | Descriptor: | MAGNESIUM ION, TPR_REGION domain-containing protein | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-17 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Thermophage RNA polymerase
To Be Published
|
|
8F5M
 
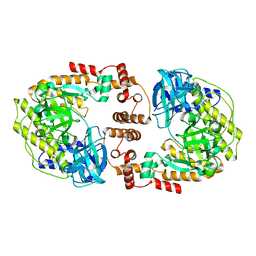 | | Crystal structure of P74 gp62 | | Descriptor: | Envelope glycoprotein gp62, MAGNESIUM ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2022-11-14 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tail-tape-fused virion and non-virion RNA polymerases of a thermophilic virus with an extremely long tail.
Nat Commun, 15, 2024
|
|
1J98
 
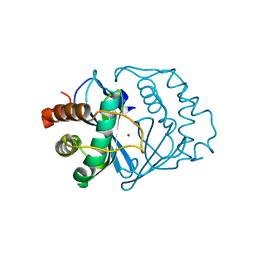 | | The 1.2 Angstrom Structure of Bacillus subtilis LuxS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A Structure of a Novel Quorum-Sensing Protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1J2X
 
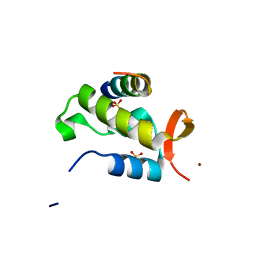 | | Crystal structure of RAP74 C-terminal domain complexed with FCP1 C-terminal peptide | | Descriptor: | RNA polymerase II CTD phosphatase, SULFATE ION, Transcription initiation factor IIF, ... | | Authors: | Kamada, K, Roeder, R.G, Burley, S.K. | | Deposit date: | 2003-01-15 | | Release date: | 2003-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of recruitment of TFIIF- associating RNA polymerase C-terminal domain phosphatase (FCP1) by transcription factor IIF
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1JQW
 
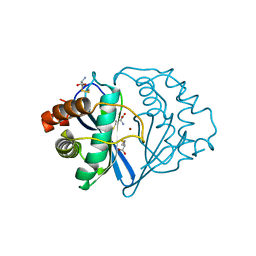 | | THE 2.3 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/HOMOCYSTEINE COMPLEX | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1JVI
 
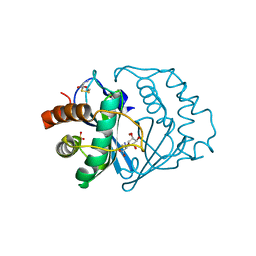 | | THE 2.2 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/RIBOSILHOMOCYSTEINE COMPLEX | | Descriptor: | (2S)-2-amino-4-[[(2S,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methylsulfanyl]butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ... | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1KBJ
 
 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: comparison with the Intact Wild-type Enzyme | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1KBI
 
 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.-W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1KEQ
 
 | | Crystal Structure of F65A/Y131C Carbonic Anhydrase V, covalently modified with 4-chloromethylimidazole | | Descriptor: | 4-METHYLIMIDAZOLE, ACETIC ACID, F65A/Y131C-MI Carbonic Anhydrase V, ... | | Authors: | Jude, K.M, Wright, S.K, Tu, C, Silverman, D.N, Viola, R.E, Christianson, D.W. | | Deposit date: | 2001-11-16 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of F65A/Y131C-methylimidazole carbonic anhydrase V reveals architectural features of an engineered proton shuttle.
Biochemistry, 41, 2002
|
|
8JOJ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate | | Descriptor: | 3-ketosteroid dehydrogenase, 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-07 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.819 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate
To Be Published
|
|
8KC6
 
 | | X-ray structure of 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase | | Authors: | Song, S.K, Lv, D.Q, Cheng, X.Y, Ma, Q.Q, Hu, Y.L, Su, Z.D. | | Deposit date: | 2023-08-06 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase
To Be Published
|
|
8KCZ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
