6WZB
 
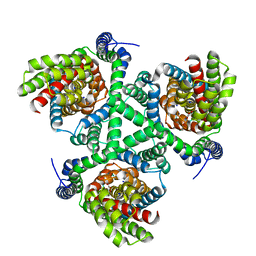 | | Crystal structure of the GltPh V216C-G388C mutant cross-linked with divalent mercury | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog, MERCURY (II) ION, ... | | Authors: | Chen, I, Font, J, Ryan, R. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
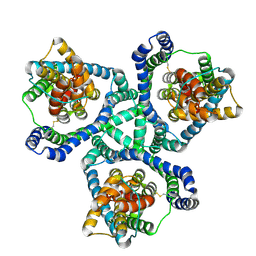
6X01
 
 | |
2NWX
 
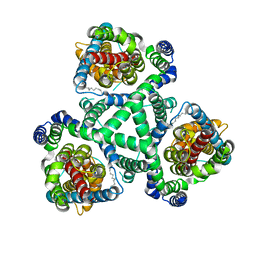 | | Crystal structure of GltPh in complex with L-aspartate and sodium ions | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, PALMITIC ACID, ... | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
2NWL
 
 | | Crystal structure of GltPh in complex with L-Asp | | Descriptor: | ASPARTIC ACID, PALMITIC ACID, glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
2NWW
 
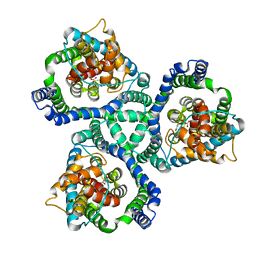 | | Crystal structure of GltPh in complex with TBOA | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, 425aa long hypothetical proton glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
7LQK
 
 | | Crystal structure of the R375A mutant of LeuT | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
7LQJ
 
 | | Crystal structure of LeuT bound to L-Alanine | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
7LQL
 
 | | Crystal structure of the R375D mutant of LeuT | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
6YIG
 
 | | Crystal structure of the N-terminal EF-hand domain of Arabidopsis thaliana AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein, SODIUM ION | | Authors: | Yperman, K, Merceron, R, De Munck, S, Bloch, Y, Savvides, S.N, Pleskot, R, Van Damme, D. | | Deposit date: | 2020-04-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YEU
 
 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YET
 
 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
5XYH
 
 | | Crystal Structure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae | | Descriptor: | CbsA | | Authors: | Kumar, S, Haque, A.S, Nathawat, R, Sankaranaryanan, R. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Mol. Plant Pathol., 19, 2018
|
|
2QXT
 
 | |
2QXU
 
 | |
