5YVO
 
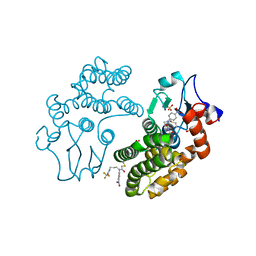 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | 分子名称: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | 著者 | Saisawang, C, Ketterman, A, Wongsantichon, J. | | 登録日 | 2017-11-27 | | 公開日 | 2018-11-28 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, M06 | | 著者 | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | 登録日 | 2014-05-20 | | 公開日 | 2014-05-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
6IM8
 
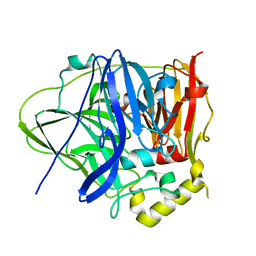 | | CueO-PM2 multicopper oxidase | | 分子名称: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO | | 著者 | Wongsantichon, J, Robinson, R, Ghadessy, F. | | 登録日 | 2018-10-22 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM9
 
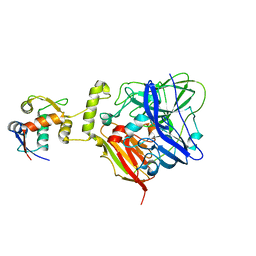 | | MDM2 bound CueO-PM2 sensor | | 分子名称: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Wongsantichon, J, Robinson, R, Ghadessy, F. | | 登録日 | 2018-10-22 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM7
 
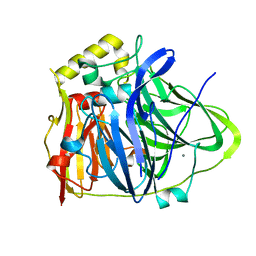 | | CueO-12.1 multicopper oxidase | | 分子名称: | Blue copper oxidase CueO,12.1 peptide,Blue copper oxidase CueO, CALCIUM ION | | 著者 | Wongsantichon, J, Robinson, R, Ghadessy, F. | | 登録日 | 2018-10-22 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
5WDH
 
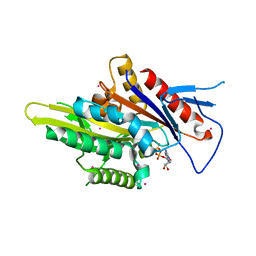 | | Motor domain of human kinesin family member C1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC1, MAGNESIUM ION, ... | | 著者 | Zhu, H, Tempel, W, He, H, Shen, Y, Wang, J, Brothers, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-07-05 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.248 Å) | | 主引用文献 | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
5WDE
 
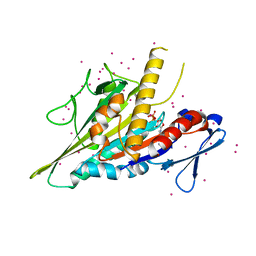 | | Crystal structure of the KIFC3 motor domain in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC3, MAGNESIUM ION, ... | | 著者 | Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-07-05 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
5XXK
 
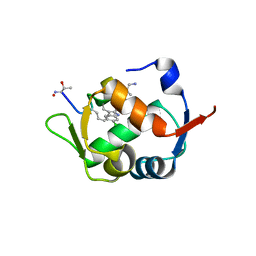 | |
6JSU
 
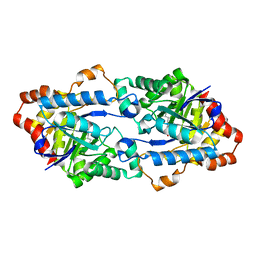 | |
6JST
 
 | |
6JSS
 
 | |
5H3N
 
 | |
5H3M
 
 | |
1D0N
 
 | | THE CRYSTAL STRUCTURE OF CALCIUM-FREE EQUINE PLASMA GELSOLIN. | | 分子名称: | HORSE PLASMA GELSOLIN | | 著者 | Burtnick, L.D, Robinson, R, Li, C. | | 登録日 | 1999-09-13 | | 公開日 | 1999-09-15 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation.
Cell(Cambridge,Mass.), 90, 1997
|
|
5YQW
 
 | | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | 著者 | Suginta, W, Sritho, N, Ranok, A, Kitaoku, Y, Bulmer, D.M, van den Berg, B, Fukamizo, T. | | 登録日 | 2017-11-08 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marineVibriobacteria.
J. Biol. Chem., 293, 2018
|
|
3JS6
 
 | |
3ARQ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with IDARUBICIN | | 分子名称: | Chitinase A, GLYCEROL, IDARUBICIN | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS3
 
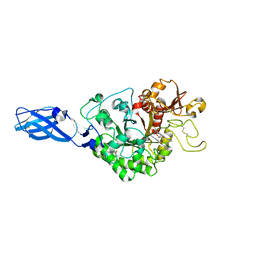 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | 分子名称: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARZ
 
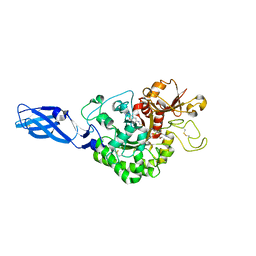 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | 分子名称: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARV
 
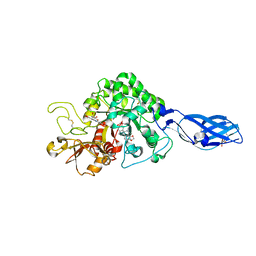 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Sanguinarine | | 分子名称: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS0
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Sanguinarine | | 分子名称: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARR
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with PENTOXIFYLLINE | | 分子名称: | 3,7-DIMETHYL-1-(5-OXOHEXYL)-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, Chitinase A | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS2
 
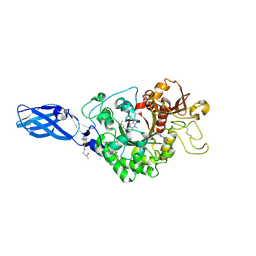 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Propentofylline | | 分子名称: | 3-methyl-1-(5-oxohexyl)-7-propyl-3,7-dihydro-1H-purine-2,6-dione, Chitinase A | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARS
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - apo structure of mutant W275G | | 分子名称: | Chitinase A | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ARU
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with PENTOXIFYLLINE | | 分子名称: | 3,7-DIMETHYL-1-(5-OXOHEXYL)-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, Chitinase A | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
