6M98
 
 | |
6M97
 
 | | Crystal structure of the high-affinity copper transporter Ctr1 | | Descriptor: | Chimera protein of High affinity copper uptake protein 1 and Soluble cytochrome b562, HEXATANTALUM DODECABROMIDE, ZINC ION | | Authors: | Ren, F, Yuan, P. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | X-ray structures of the high-affinity copper transporter Ctr1.
Nat Commun, 10, 2019
|
|
3W02
 
 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W01
 
 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VKD
 
 | | Crystal structure of MoeO5 soaked with 3-phosphoglycerate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VK5
 
 | | Crystal structure of MoeO5 in complex with its product FPG | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-08 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKC
 
 | | Crystal structure of MoeO5 soaked with pyrophosphate | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VZZ
 
 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZX
 
 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W00
 
 | | Crystal structure of PcrB complexed with G1P and FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VKB
 
 | | Crystal structure of MoeO5 soaked with FsPP overnight | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MoeO5, ... | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3VKA
 
 | | Crystal structure of MoeO5 soaked for 3 hours in FsPP | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6HQG
 
 | | Cytochrome P450-153 from Phenylobacterium zucineum | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQW
 
 | |
5AMS
 
 | | Crystal structure of Sqt1 | | Descriptor: | RIBOSOME ASSEMBLY PROTEIN SQT1 | | Authors: | Frenois, F, Legrand, P, Fribourg, S. | | Deposit date: | 2015-08-31 | | Release date: | 2016-01-13 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Sqt1P is an Eight-Bladed Wd40 Protein
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2VY6
 
 | | Two domains from the C-terminal region of influenza A virus polymerase PB2 subunit | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
2VY7
 
 | | The 627-domain from influenza A virus polymerase PB2 subunit | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-09 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
2VY8
 
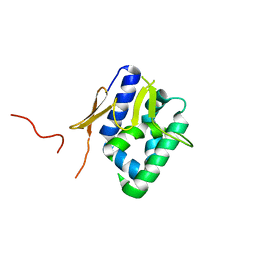 | | The 627-domain from influenza A virus polymerase PB2 subunit with Glu- 627 | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
2JDQ
 
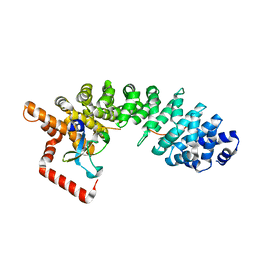 | | C-terminal domain of influenza A virus polymerase PB2 subunit in complex with human importin alpha5 | | Descriptor: | IMPORTIN ALPHA-1 SUBUNIT, POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Guilligay, D, Mas, P, Boulo, S, Baudin, F, Ruigrok, R.W.H, Hart, D.J, Cusack, S. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Nuclear Import Function of the C- Terminal Domain of Influenza Virus Polymerase Pb2 Subunit
Nat.Struct.Mol.Biol., 14, 2007
|
|
1U9N
 
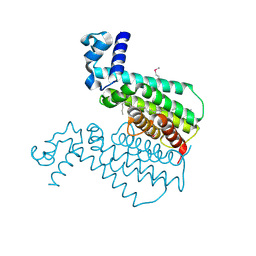 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation opens therapeutic perspectives against tuberculosis and leprosy | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
1U9O
 
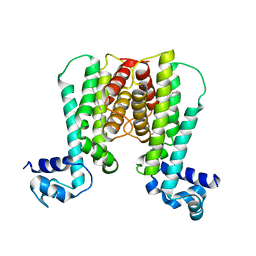 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
2C1W
 
 | | The structure of XendoU: a splicing independent snoRNA processing endoribonuclease | | Descriptor: | ENDOU PROTEIN, PHOSPHATE ION | | Authors: | Renzi, F, Caffarelli, E, Laneve, P, Bozzoni, I, Brunori, M, Vallone, B. | | Deposit date: | 2005-09-21 | | Release date: | 2006-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Endoribonuclease Xendou: From Small Nucleolar RNA Processing to Severe Acute Respiratory Syndrome Coronavirus Replication.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7QH5
 
 | |
