6VYH
 
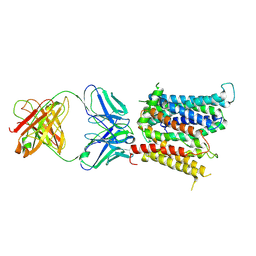 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
3NOU
 
 | |
3NOP
 
 | |
3NOT
 
 | |
3HFH
 
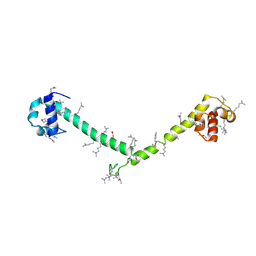 | | Crystal structure of tandem FF domains | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Lu, M, Yang, J, Ren, Z, Subir, S, Bedford, M.T, Jacobson, R.H, McMurray, J.S, Chen, X. | | Deposit date: | 2009-05-11 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of the Three Tandem FF Domains of the Transcription Elongation Regulator CA150.
J.Mol.Biol., 393, 2009
|
|
4RO0
 
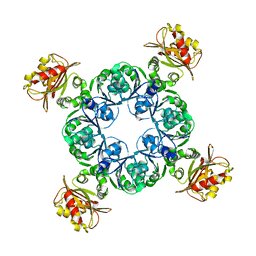 | | Crystal structure of MthK gating ring in a ligand-free form | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Dong, W, Guo, R, Chai, H, Chen, Z, Cui, H, Ren, Z, Li, Y, Ye, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The principle of cooperative gating in a calcium-gated K+ channel
To be Published
|
|
6UVB
 
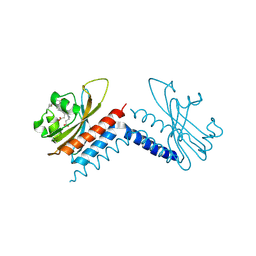 | |
6UV8
 
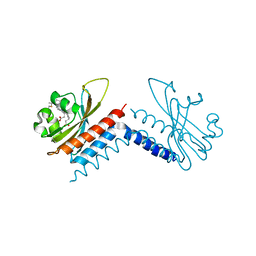 | |
6VP0
 
 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
5TUW
 
 | | Crystal structure of Orange Carotenoid Protein with partial loss of 3'OH Echinenone chromophore | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TV0
 
 | | crystal structure and light induced structural changes in orange carotenoid protein bound with 3 'OH echinenone | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, GLYCEROL, Orange carotenoid-binding protein | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7TAK
 
 | | Structure of a NAT transporter | | Descriptor: | GUANINE, Putative membrane protein PurT | | Authors: | Weng, J, Zhou, X, Ren, Z, Chen, K, Zhou, M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79828 Å) | | Cite: | Structure of a NAT transporter
To Be Published
|
|
5TUX
 
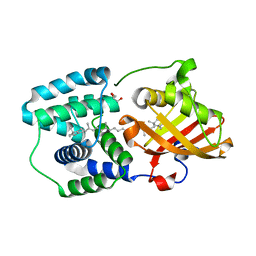 | | crystal structure and light induced structural changes in orange carotenoid protein bound with echinenone | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3NHQ
 
 | | The dark Pfr structure of the photosensory core module of P. aeruginosa Bacteriophytochrome | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Yang, X, Ren, Z, Kuk, J, Moffat, K. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome.
Nature, 479, 2011
|
|
5IWS
 
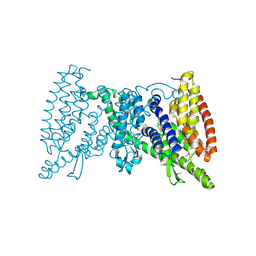 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
4NAA
 
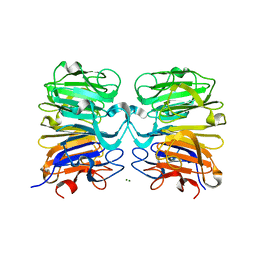 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | Deposit date: | 2013-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NC4
 
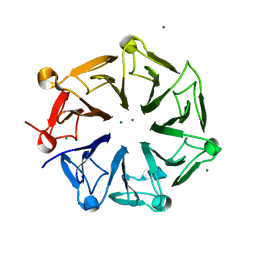 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
3RTP
 
 | | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration | | Descriptor: | Mitogen-activated protein kinase 10, N-[4-cyano-3-(1H-1,2,4-triazol-5-yl)thiophen-2-yl]-2-(2-oxo-3,4-dihydroquinolin-1(2H)-yl)acetamide | | Authors: | Bowers, S, Truong, A.P, Neitz, R.J, Hom, R.K, Sealy, J.M, Probst, G.D, Quincy, Q, Peterson, B, Chan, W, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Pan, H, Lin, M, Yao, N, Artis, D.R, Zhang, H, Chen, L, Dryer, M, Samant, B, Zmolek, W, Wong, K, Lorentzen, C, Goldbach, E, Tonn, G, Quinn, K.P, Sauer, J, Wright, S, Powell, K, Ruslim, L, Ren, Z, Bard, F, Yednock, T.A, Griswold-Prenne, I. | | Deposit date: | 2011-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
430D
 
 | | STRUCTURE OF SARCIN/RICIN LOOP FROM RAT 28S RRNA | | Descriptor: | MAGNESIUM ION, SARCIN/RICIN LOOP FROM RAT 28S R-RNA | | Authors: | Correll, C.C, Munishkin, A, Chan, Y.L, Ren, Z, Wool, I.G, Steitz, T.A. | | Deposit date: | 1998-10-04 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ribosomal RNA domain essential for binding elongation factors.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4I5M
 
 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4IFJ
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
4IFN
 
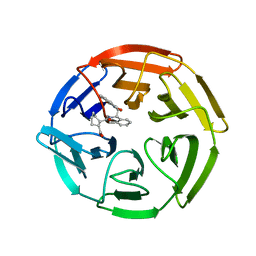 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4IFL
 
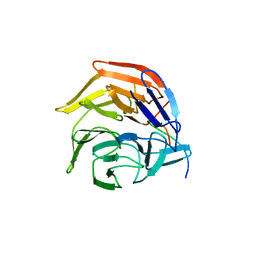 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | Descriptor: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | Authors: | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4NBM
 
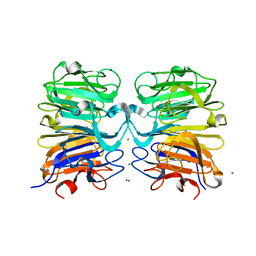 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
7UKN
 
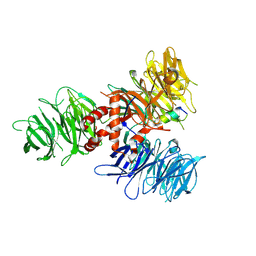 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of pUL145 | | Descriptor: | DNA damage-binding protein 1, H-Box Motif of pUL145 | | Authors: | Wick, E.T, Treadway, C.J, Nicely, N.I, Li, Z, Ren, Z, Baldwin, A.S, Xiong, Y, Harrison, J.S, Brown, N.G. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into Viral Hijacking of CRL4 Ubiquitin Ligase through Structural Analysis of the pUL145-DDB1 Complex.
J.Virol., 96, 2022
|
|
